Figure 5.
Amino Acid Signatures Are Informative about Gene Function and Provide Orthogonal Information to Other Molecular Networks
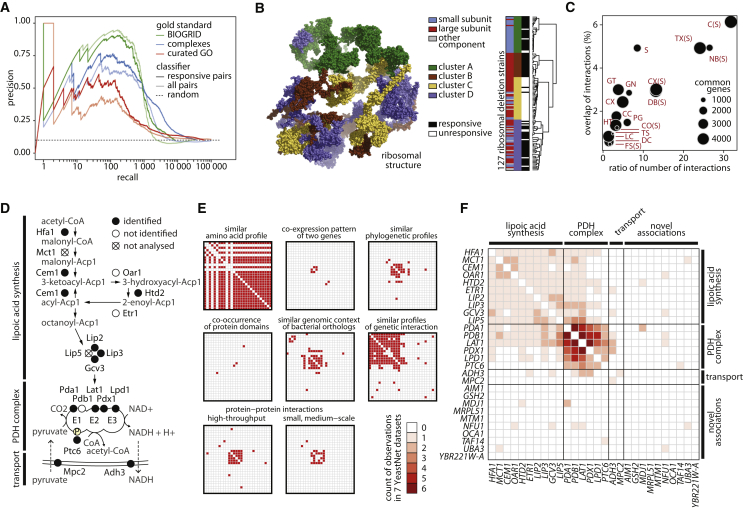
(A) Precision-recall analysis evaluating metabolite profiles in comparison to curated protein complexes, low-throughput genetic and physical protein-protein interactions, and GO terms. The different gold standards are color coded and the set of metabolic signatures distinguished by transparency; the dashed line corresponds to the random classifier.
(B) Metabolic signatures reflect structural proximity of the 127 non-essential ribosomal proteins, mapped to the ribosomal structure (PDB: 4V8Y) (Fernández et al., 2013).
(C) Orthogonality of functional metabolomics to other molecular data. Metabolic signatures were assembled in a phenotypic association network and compared to STRING and YeastNet functional genomic datasets. y axis is the overlap of edges between the genes (nodes) present in the compared networks, whereas the x axis is the ratio of edges, comparing the STRING or YeastNet, to the metabolic clustering. Circle size indicates the number of genes compared. Abbreviations as in YeastNet (given in full in Figure S4C). Metabolic signatures provide highly orthogonal information, and the best agreement is obtained upon combining all data as contained in STRING (C(S)).
(D) Metabolic signatures provide orthogonal information to existing molecular networks, as exemplified by the lipoic acid (LA) biosynthesis pathway, which provides the cofactor for the pyruvate dehydrogenase (PDH) complex. Scheme of the pathway is shown.
(E) Association of metabolic signatures (LA and PDH cluster), in comparison to yeast genetic and physical interaction networks as accessed through YeastNet. A majority of the associations among the LA biosynthetic genes are obtained only over the metabolic signature.
(F) Summarizing the information content of all molecular networks (E), except metabolic signature, in comparison to the association obtained over the metabolic signature (full matrix). When combined, genetic and physical interaction networks capture the PDH complex but provide only partial coverage of the LA pathway.