Figure S4.
Metabolic Signatures Add New Functional Information to Incompletely Characterized S. cerevisiae Genomes, Related to Figure 5
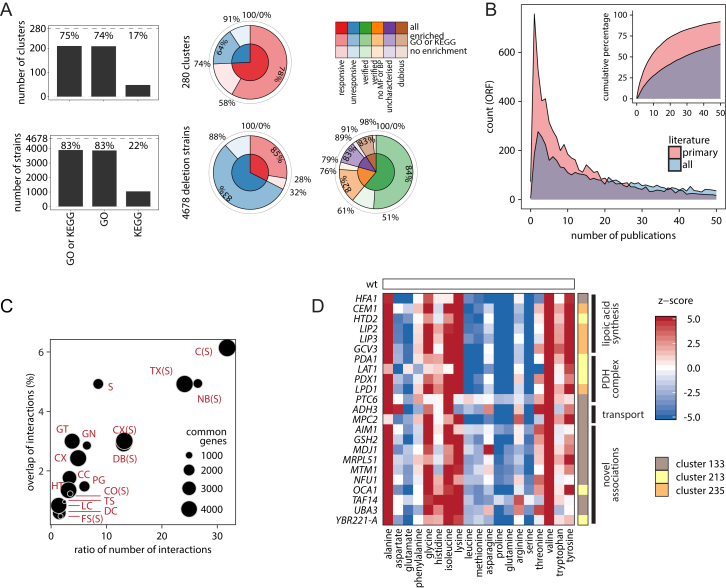
(A) (top, left panel) Functional enrichment analysis in 280 clusters for GO and KEGG terms. Enrichment was comparable in clusters with at least one significantly changed amino acid (responsive) or in clusters without (unresponsive), reflecting that small co-occurring amino acid concentration changes reveal functionally relevant signature (top, middle panel); functional enrichment analysis as described in STAR Methods. For 83% of gene deletions strains, a functional association was predicted. (bottom panel) 39% percent of deletion strains are not well characterized according to our definition (no GO or function or process term associated, annotated as ‘uncharacterized’). A functional association is possible for 83% of them.
(B) To quantitatively enumerate the occurrence of so far poorly and uncharacterized genes in budding yeast, we orient on the most comprehensive collection of functional information in budding yeast, the Saccharomyces genome database, which combines large-scale, small scale, literature mining and community-driven approaches (Cherry et al., 2012). Primary literature contains information about “function, biological role, cellular location, phenotype, regulation, structure, or disease homologs in other species for the gene or gene product.” All literature further incorporates reviews and publications with “experimental evidence for the gene or describe homologs in other species, but for which the gene is not the paper’s principal focus.” 26.7% of yeast ORFs have so far been analyzed in only three to zero dedicated studies, 22% of open reading frames in SGD are considered functionally fully uncharacterized according to systematic annotation, while 35.8% lack so far a gene ontology function or process term association. Genes that are annotated as uncharacterized or lack a GO annotation sum up to 1823 (39%) of the strains analyzed in our study.
Orthogonality of functional metabolomics to other molecular data. Abbreviations are as described in YeastNet resource, for every network links are inferred from a different source, as following: CC - co-citation of two genes across 46,111 PubMed Medline article abstracts for yeast biology; CX - co-expression pattern of two genes (based on high-dimensional gene expression data); DC - co-occurrence of protein domains between two coding genes; GN - similar genomic context of bacterial orthologs of two yeast genes; GT - similar profiles of genetic interaction partner; HT - high-throughput protein-protein interactions; LC - small/medium-scale protein-protein interactions (collected from protein-protein interaction databases); PG - similar phylogenetic profiles between two yeast genes; TS - 3-D protein structure of interacting orthologous proteins between two yeast proteins. The rest interaction networks based on evidences collected in STRING database, i.e., DB(S) - based on pathways from curated databases; NB(S) - interactions inferred from genomic neighbors; CX(S) - interactions inferred using gene co-expression; CO(S) - gene co-occurrence in phylogenetic trees; TX(S) - interactions based on automated unsupervised text mining when searching for proteins that are frequently mentioned together; F(S) - genes that are sometimes fused together; C(S) - based on all combined evidences in STRING.
(C) Perturbation of lipoic acid synthesis or the pyruvate dehydrogenase reaction triggers a highly similar amino acid profile and allows to associate a novel gene function to cluster members. Clusters 133, 213 and 235 are joined at greater branch height and form a cluster separated from the remaining deletion strain profiles (Data S1). The deleted ORF
GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological process; MF, molecular function.