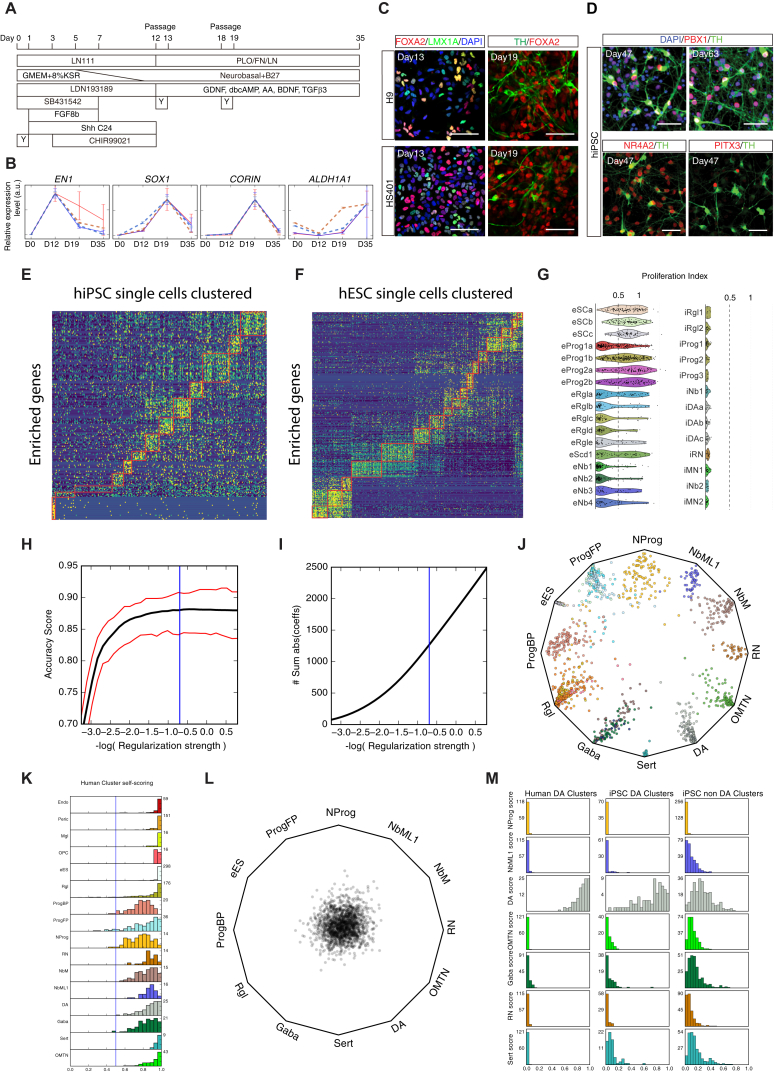
Figure S7.
Stem Cells Differentiation Protocol and Machine Learning Performance and Diagnostics, Related to Figure 7
(A) Schematic of the hESCs in vitro differentiation protocol.
(B) Variation of expression of marker genes during the differentiation protocol measured by qPCR. As a comparison, values calculated by summing single cell expression levels are shown as dashed line.
(C) Immunostaining of hESC cultures (scale bar 50 μm).
(D) Immunostaining of hiPSC cultures (scale bar 50 μm).
(E) Heatmap showing raw hiPSCs data clustered. Columns are single cell and rows genes. Cell clusters and genes enriched in every cluster are surrounded by red boxes.
(F) Heatmap showing raw hESCs data as in (E).
(G) Violin plots showing proliferation index distribution for each hiPSC and hESC cluster.
(H) Line plot showing the accuracy score of the classifier varying with decreasing regularization strength as estimated by cross-validation. Red line shows 95% C.I. on the estimation of the accuracy score.
(I) Line plot showing the total absolute values of weights. Blue vertical line in (H-I) shows the chosen regularization parameter.
(J) Training dataset plotted on wheel plot as in Figure 7G.
(K) Score distribution on the training dataset clusters.
(L) Negative control cells are obtained by scrambling gene values of cells of the training dataset and are plotted on wheel plot as in Figure 7G.
(M) Histograms showing classifier scores of single cells belonging to the human dopaminergic clusters (left), to the dopaminergic hiPSC clusters (center) and to the rest of the cells in the hiPSC preparation (right).