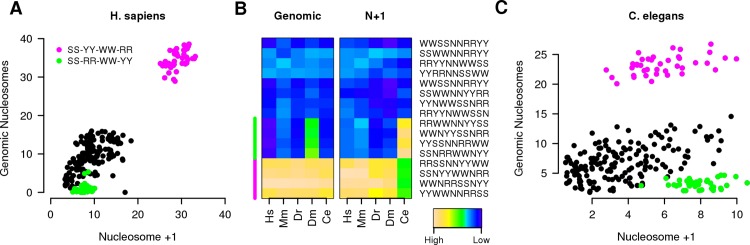
Fig 2. Identification of the N+1 consensus sequence.
(A) Correlation between 10 bp long dinucleotide patterns composed of one copy of each SS, WW, YY and RR dinucleotides and 2 Ns evaluated on human genomic nucleosomes and the N+1 nucleosome; each dot represents the strength of the 10 bp frequency of a consensus sequence in the N+1 region or in genomic nucleosome regions (defined by MNase-seq data); green and red dots mark two classes of patterns (circular permutations of SS-RR-WW-YY and SS-YY-WW-RR, respectively) that are known to have high nucleosomes affinities [47]. (B) Comparative analysis of representative consensus sequence strength in 5 organisms on genomic nucleosomes (left panel, Genomic) and promoter nucleosomes (right panel, N+1). Each square represents the strength of a 10 bp periodic signal for a consensus sequence (rows) for an organism (columns). Hs: H. sapiens, Mm: M. musculus, Dr: D. rerio, Dm: D. melanogaster and Ce: C. elegans. A comparative analysis of all motifs tested can be found in S6 and S7 Figs (C) Correlation analysis as performed in A for C. elegans promoters and genomic nucleosomes.