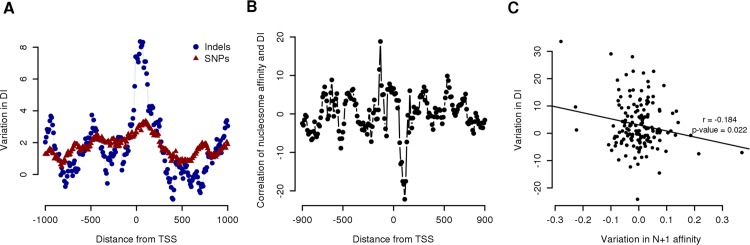
Fig 4. Effects of promoter natural variants on Pol-II initiation.
(A) Analysis of the effect of SNPs or Indels to Pol-II initiation precision as a function of their distance from the TSS. Values on the x axis represent mid point of a sliding window of 150 bp (shift 10 bp) whereas y axis represent the variation in DI between GM12878 cell line and other blood derived cell lines for promoters that have natural variants in the GM12878 cell line mapping in that region. (B) 2 Kb region around human promoters were scanned for the presence of natural variants with a sliding window of 150 bp (shift 10) and analyzed for their impact on WW 10 bp frequency intensity in that region. The variation in signal intensity was then correlated to variation in DI for the corresponding promoters using a linear model. The dots represent the slope (angular coefficient) of the linear model for the region centered in that position. Negative slope values correspond to negative correlation between the variation in WW dinucleotide frequency and variation in DI (C) Effects of mutations in the N+1 region on nucleosome affinity and DI. The plot shows the effect of mutation on nucleosome affinity in the N+1 region (measured as the difference in 10 bp frequency intensity for WW dinucleotide between GM12878 and ML sequences) and its correlation with variation in DI for the same promoter (measured as difference between GM12878 and other blood related cell lines). Each dot represents a different promoter. The p-value of 0.022 corresponds to the F statistic evaluated on the linear regression model.