Aligned carbon nanotube–integrated device can effectively trap and enrich viruses from field samples without using antibodies.
Keywords: Virus, carbon nanotubes, N-doped, influenza, enrichment, field samples, sensor, improved detection, NGS, PCR
Abstract
Viral infectious diseases can erupt unpredictably, spread rapidly, and ravage mass populations. Although established methods, such as polymerase chain reaction, virus isolation, and next-generation sequencing have been used to detect viruses, field samples with low virus count pose major challenges in virus surveillance and discovery. We report a unique carbon nanotube size-tunable enrichment microdevice (CNT-STEM) that efficiently enriches and concentrates viruses collected from field samples. The channel sidewall in the microdevice was made by growing arrays of vertically aligned nitrogen-doped multiwalled CNTs, where the intertubular distance between CNTs could be engineered in the range of 17 to 325 nm to accurately match the size of different viruses. The CNT-STEM significantly improves detection limits and virus isolation rates by at least 100 times. Using this device, we successfully identified an emerging avian influenza virus strain [A/duck/PA/02099/2012(H11N9)] and a novel virus strain (IBDV/turkey/PA/00924/14). Our unique method demonstrates the early detection of emerging viruses and the discovery of new viruses directly from field samples, thus creating a universal platform for effectively remediating viral infectious diseases.
INTRODUCTION
Viruses may cause unpredictable and recurring outbreaks that lead to devastating mortality and traumatic economic losses, as exemplified by the 1918 influenza pandemic, the ongoing battle against HIV/AIDS, and the most recent Ebola and Zika outbreaks (1, 2). However, there is still a large pool of unknown mammalian and human viruses among which could be critical viral pathogens (3, 4). Almost all lethal viral outbreaks in the past two decades were caused by newly emerging viruses (5). Because more than 50% of the human pathogens are known to be zoonotic (6, 7), virus samples can be originated from various sources, for example, humans, animals, and different environments. Thus, it is clear that the successful virus isolation, identification, and genome characterization, directly from field and clinical samples, will lead to rapid discovery of emerging viral pathogens (8).
Because the high mutation rate and the genetic diversity of viruses warrant extensive surveillance (9), various virus detection approaches have been established: (i) enzyme-linked immunosorbent assay (ELISA) (10), (ii) polymerase chain reaction (PCR) (11, 12), (iii) virus isolation (13, 14), and (iv) next-generation sequencing (NGS) (5, 15). However, additional advancements in the sample preparation techniques are urgently needed to enrich and concentrate viruses (16–20). In addition, the most conventional virus sample preparation protocols use immunological capture, physical separation, or a combination of both (21, 22). Unfortunately, immunological capture requires previous knowledge of the targets; thus, it is not appropriate for virus discovery and can lead to technical difficulties in identifying new or emerging virus strains. Ultracentrifugation is the most commonly used physical method for virus enrichment and concentration. Unfortunately, it involves bulky equipment, intensive labor, and lengthy sample preparation, and has limitations for concentrating small amounts of viruses in minute volumes (15, 22, 23). Microfiltration membranes can remove large particles within samples while keeping the virus particles in the supernatant. It is normally used as one of the steps in the whole sample preparation protocol for virus analysis; however, it neither removes contaminants of small size (for example, nucleic acids and proteins) nor concentrates the sample (24–26). Although ultrafiltration membranes are widely used as an essential viral clearance step in the biopharmaceutical production from human or animal origin (21, 27), their usage for virus detection is rare, primarily because of their low porosity, high operation pressure, poor virus viability, and difficulty in virus access for further analysis.
In this context, robust arrays of aligned carbon nanotubes (CNTs) with controlled intertube distance could be used to effectively trap/concentrate viruses within a three-dimensional (3D) porous system. Although CNTs have been used as biochemical sensors (28), imaging probes (29), drug delivery vehicles (30), x-ray sources (31), neuron protection (32), treatment of drug addiction (33), and substrates for immunological capture of mammalian cells and bacteria (34), they have not been integrated into tunable devices able to isolate viruses of different sizes. Here, we developed a reliable, scalable CNT size-tunable enrichment microdevice (CNT-STEM) technology that provides size-based, label-free, viable enrichment of viruses from field samples. We synergistically integrated bottom-up controlled nanotube synthesis with top-down microfabrication. We demonstrated that the CNT-STEM not only enriches viruses from field samples by at least 100 times (Fig. 1) but also removes host and environmental contaminants and concentrates samples to enable direct virus identification by NGS from field-collected samples.
Fig. 1. The working principle of virus enrichment and concentration from field samples.
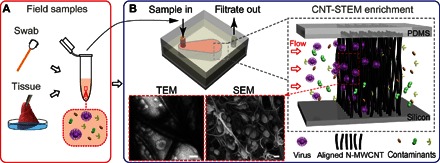
(A) A field sample containing viruses (purple spheres) is collected by a cotton swab or as a tissue sample. (B) The supernatant of the field sample flows through the CNT-STEM, and the viruses are enriched within the device. Inset (right): Illustration of size-based virus enrichment by the aligned N-MWCNTs. Inset (bottom right): SEM image (scale bar, 100 nm) of the H5N2 AIV virions trapped inside the aligned N-MWCNTs. Inset (bottom left): Dark-field TEM image (scale bar, 100 nm) of enriched H5N2 AIV after the aligned N-MWCNTs structures were retrieved from the CNT-STEM.
RESULTS
Tunable intertubular distance of aligned N-MWCNTs
Depending on the type and source of the virus-containing sample, virus particles need to be released into aqueous suspensions by gentle vortexing, shaking (for swab samples), or tissue homogenization (for tissue samples) before they are introduced into the CNT-STEM (Fig. 1A). We normally remove large cells or tissue debris by filtering the crude samples with membrane filters exhibiting pore size of 220 or 450 nm. As the virus suspension flows through the CNT-STEM, virus particles are efficiently captured by the nitrogen-doped multiwalled CNT (N-MWCNT) arrays while contaminants of small size flow through (Fig. 1B). If needed, the viruses (tightly adhered to the CNTs) can be easily retrieved/studied after opening the device (fig. S1). To synthesize these vertically aligned N-MWCNTs on the microdevice substrate, we used standard semiconductor batch microfabrication techniques to pattern catalyst (iron clusters), followed by selective growth using aerosol-assisted chemical vapor deposition (AACVD; Fig. 2A and fig. S2) (35, 36).
Fig. 2. Controlled N-MWCNT growth and tunable size-based filtration of the CNT-STEM.
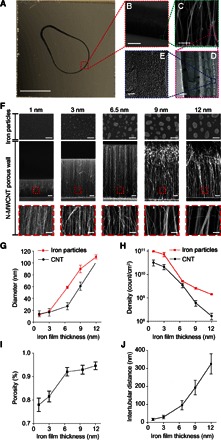
Microscopy images of the N-MWCNT porous wall in the CNT-STEM (A to E). (A) Bright-field optical microscopy image showing the top view of the droplet-shaped porous wall made by the aligned N-MWCNTs on a silicon substrate. Scale bar, 2 mm. (B) SEM image of the aligned N-MWCNTs. Scale bar, 50 μm. (C) High-magnification SEM image showing a side view of the aligned N-MWCNTs. Scale bar, 100 nm. (D) TEM image of AACVD synthesized N-MWCNTs of various diameters. Scale bar, 20 nm. (E) High-resolution TEM image showing the multiwall structure N-MWCNT. Scale bar, 5 nm. (F) Formation of iron nanoparticle catalyst and growth of N-MWCNTs on iron catalyst layers with different thicknesses. Top row: SEM images showing top views of iron particles formed on a silicon surface after 850°C thermal treatment in AACVD. Thicknesses of iron catalyst thin films are 1, 3, 6.5, 9, and 12 nm. Middle and bottom row: SEM images of cross-sectional views of an aligned N-MWCNT structure after 30-min N-MWCNT growth by AACVD. Scale bars, 100 nm (top), 10 μm (middle), 200 nm (bottom). Diameter (G) and density (H) of iron particles (red) and N-MWCNT (black) as a function of iron film thickness (n = 8). (I) Intertubular distance measured by image analysis as a function of iron film thickness (n = 8). (J) Calculated porosity of the N-MWCNT wall (n = 8).
Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) images (Fig. 2, B to E) as well as Raman measurements with calculated D/G intensity ratios (fig. S3, A and B) confirm the presence of N-MWCNTs synthesized directly on the substrates (35–37). N-MWCNTs were selected for their excellent mechanical strength (38) and optimal biocompatibility as reported in our previous study (39, 40). After N-MWCNT arrays were grown on patterned areas of the substrate, we bonded the silicon substrate with a polydimethylsiloxane (PDMS) chamber to perfectly seal the microfluidic chamber without experiencing any leakage.
An important accomplishment of this work is the control of the intertubular distance within the CNT arrays so they could match different virus sizes. In this context, different iron catalyst thicknesses were deposited onto the Si substrates (Fig. 2, F to J). When the thickness of the iron catalyst thin film increases from 1 to 12 nm, the density of the iron particles decreases while the particle diameter increases, thus causing the intertubular distance of N-MWCNTs to increase from 17 ± 6 to 325 ± 56 nm. It is also noteworthy that N-MWCNTs consist of concentric tubules exhibiting average diameters of 17 to 99 nm. In general, the height of the N-MWCNTs also increases over time; however, the growth rate significantly decreases after 20 to 30 min of synthesis (fig. S3, C to F).
Performance of size-tunable enrichment characterization
To validate the size-tunable enrichment capability of the CNT-STEM, we tested and introduced fluorescent molecules and fluorescent polystyrene nanospheres of 20, 50, 100, 140, 400, and 1000 nm in diameter into CNT-STEMs exhibiting different intertubular distances (Fig. 3A and fig. S4A). Figure 3B shows the filtration characteristics of CNT-STEMs with 25-, 95-, and 325-nm intertubular distances. They all have a binary separation profile, which means that for a CNT-STEM with a particular intertubular distance, smaller nanoscale particles usually penetrate the N-MWCNT structure, whereas larger particles cannot. We defined the particle diameter corresponding to a 50% penetration ratio (the background-corrected fluorescence intensity of the filtrate to that of the original suspension) as the critical particle size of the CNT-STEM with a specific intertubular distance (table S1). However, the fluorescence intensity inside the N-MWCNT array is extremely low, maintaining at the same level before and after fluorescein or fluorescence nanospheres flow into the device. This can be explained by the high optical absorbance of the N-MWCNT forest, reported for vertically aligned CNT forests as a nearly perfect blackbody absorber (41–43). Similarly, the viruses inside the N-MWCNT array also elude fluorescence detection (Fig. 4A).
Fig. 3. Measured particle size–dependent filtration characteristics of CNT-STEMs with N-MWCNT intertubular distances of 25, 95, and 325 nm, using small-molecule fluorescein and fluorescent polystyrene nanospheres of 20, 50, 100, 140, 400, and 1000 nm in diameter.

(A) Fluorescence microscopy images showing fluorescein solution and fluorescent polystyrene nanospheres of various diameters being filtered by the CNT-STEM. The direction of the flow is from right to left as indicated by the red arrows. Yellow lines delineate the contours of the N-MWCNT structures. Scale bars, 50 μm. (B) Penetration of fluorescein and fluorescence polystyrene nanospheres through the N-MWCNT structure (n = 8).
Fig. 4. Enrichment and concentration of virus swab samples by CNT-STEM.
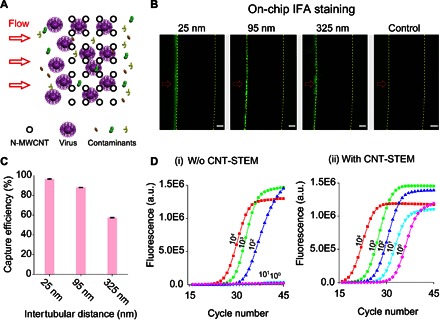
(A) Top-view illustration of viruses passing through and captured by the N-MWCNT array. (B) On-chip IFA staining of captured H5N2 AIV inside CNT-STEMs with 25-, 95-, and 325-nm intertubular distances. Fluorescence microscopy images of the CNT-STEMs. Red arrows indicate the flow of direction. Yellow dotted lines delineate the contours of the N-MWCNT structures. The control sample was allantoic fluid without viruses. Scale bars, 25 μm. (C) Capture efficiency of CNT-STEMs with intertubular distances of 25, 95, and 325 nm measured by rRT-PCR (n = 6). (D) Examples of rRT-PCR AIV detection curves for virus titers of 104, 103, 102, 101, and 100 EID50/ml without (i) and with (ii) CNT-STEM enrichment. a.u., arbitrary units.
By opening the CNT-STEM device and after observing the N-MWCNT array under SEM, we could clearly visualize the nanospheres embedded inside the N-MWCNT array (fig. S4B). Thus, to separate large nanoscale particles from small contaminates, we can tune the intertubular distance of the N-MWCNT to be smaller than the target nanoscale particles but larger than the contaminants.
Label-free capture of viruses by CNT-STEM
We used a low pathogenic (LP) avian influenza virus (AIV) (44–46) as a model system to characterize and optimize the CNT-STEM performance. In particular, we studied the performance of the CNT-STEM using swab samples of an LPAIV subtype H5N2 (A/chicken/PA/7659/1985) by spiking freshly propagated viruses into tracheal swabs obtained from specific pathogen–free (SPF) chickens. The size of the H5N2 LPAIV was measured as 93 ± 35 nm (fig. S5). When we introduced 50 μl of processed swab supernatant containing H5N2 LPAIV [107 EID50 (50% embryo infectious doses)/ml] into CNT-STEMs of 95-nm intertubular distance, SEM and TEM images clearly showed virus particles well distributed and efficiently trapped inside the N-MWCNT array (Fig. 1B, insets). The CNT-STEM–captured viruses are readily detected by on-chip indirect fluorescent antibody (IFA) assay using AIV H5 subtype–specific monoclonal antibody (Fig. 4, A and B) (47). In general, CNT-STEMs of smaller intertubular distance showed stronger fluorescence, thus indicating a higher density of the captured virus. However, as explained above, viruses trapped inside the N-MWCNT structures cannot generate fluorescence. Thus, the on-chip fluorescence staining can only qualitatively detect the existence of the viruses but is incapable of quantifying virus counts within CNT forests.
To measure virus capture efficiency, we applied conventional real-time reverse transcription PCR (rRT-PCR). We made CNT-STEMs with three different intertubular distances of 25, 95, and 325 nm. Each CNT-STEM was loaded with 50 μl of sample containing H5N2 LPAIV (106 EID50/ml). By measuring the original virus titer and that of the flow-through after enrichment with CNT-STEM, virus capture efficiency of the CNT-STEMs with 25-, 95-, and 325-nm intertubular distances was measured as 96.5 ± 0.5%, 88.0 ± 0.3%, and 57.5 ± 0.4%, respectively (Fig. 4C and figs. S6 and S7).
Virus concentration and enrichment
The most commonly used viral surveillance tests are rRT-PCR (11) and virus isolation (13, 14), where a major challenge is to yield true-positive results for samples containing virus concentrations below the detection limits. We investigated how our CNT-STEM could enrich virus sample for rRT-PCR and virus isolation. For further disease or virology study, captured viruses need to be retrieved from the device. In our case, this has been easily achieved by opening the PDMS chamber of the device and recovering the virus embedded within N-MWCNTs using a pipette tip.
To investigate the benefit of our CNT-STEM on the overall rRT-PCR assay sensitivity, we loaded 1.0 ml of H5N2 sample into CNT-STEMs of 25-nm intertubular distance. The viruses were enriched, retrieved, and resuspended in viral transport medium (UTM, catalog no. 331C) with a final volume of 50 μl. The same volume was used for conventional rRT-PCR without virus enrichment. After the CNT-STEM enrichment, rRT-PCR detected AIV in all samples (6 of 6) with original titer as low as 1 EID50/ml, whereas without using the CNT-STEM, the rRT-PCR detection limit was measured as 102 EID50/ml for the same AIV samples (Fig. 4D and figs. S6 and S8). Therefore, after CNT-STEM enrichment, original virus samples with virus titer of at least two orders of magnitude below detection limit of standard rRT-PCR became detectable by rRT-PCR. To exclude the potential effect of N-MWCNT in rRT-PCR, we added the same amount of N-MWCNTs inside the CNT-STEM into the rRT-PCR and found that the N-MWCNTs do not exhibit adverse effects (fig. S9).
Virus isolation remains the “gold standard” for AIV diagnostics (13). For this procedure, viable intact virus particles are inoculated into an embryonated chicken egg (ECE) and kept under proper conditions for virus cultivation. This procedure fails when the original virus concentration is too low or the viruses are nonviable or nonproliferable. Therefore, we investigated whether CNT-STEM–enriched virus samples can be directly used for virus isolation to study whether the trapped viruses are viable and then whether the enrichment procedure can potentially improve the well-established virus isolation procedure (Fig. 5A). In this context, we prepared the H5N2 AIV in three serial dilutions in titers of 104, 103, and 102 EID50/ml. After 72 hours of postinoculation in ECEs, we collected viruses from the allantoic fluid and applied a Dot-ELISA assay using antibody against AIV H5 antigen to test for the existence of viruses (Fig. 5B). The successful virus isolation rates were measured as 100, 100, and 90% for CNT-STEM–processed samples of original virus titers of 104, 103, and 102 EID50/ml, respectively (Fig. 5C). For those samples without CNT-STEM preparation, the corresponding virus isolation rates were determined as 100, 50, and 0%, respectively. Therefore, the CNT-STEM retains the virus viability and significantly improves the virus isolation rate, whereas the N-MWCNTs do not interfere with the virus cultivation process.
Fig. 5. CNT-STEM enriches viruses viably and improves the minimal virus concentration of virus isolation.
(A) Illustration showing inoculation of virus-embedded N-MWCNT structure into ECE. (B) Dot-ELISA detection of H5N2 AIV after virus cultivation in ECEs. Virus samples inoculated into chicken eggs were either from original virus samples in the titers of 102, 103, and 104 EID50/ml or CNT-STEM–enriched samples of the same corresponding original titers. A darkened spot with a positive sign indicated that H5N2 AIV successfully propagated inside the chicken egg. (C) Success rates of AIV isolation via egg inoculation with and without using the CNT-STEM for original virus titers of 104, 103, and 102 EID50/ml (n = 10). ***P ≤ 0.001.
Unknown virus enrichment and detection by NGS
Although NGS does not require previous knowledge of pathogens, the combination of CNT-STEMs for virus enrichment and NGS for virus identification can be a unique and powerful approach to discover unknown/emerging viruses. Normally, NGS requires starting genetic materials in microgram range with high purity in a small volume of tens of microliters (48), which is prohibitive for field samples of low virus count and that are highly contaminated. To explore the feasibility and develop a practical pipeline of the CNT-STEM for these field conditions, we used the H5N2 LPAIV strain that we had been testing with to prepare mimic field samples. Although this is an AIV strain isolated in 1985, its whole genome has not been sequenced before. We spiked freshly propagated viruses into tracheal swabs obtained from SPF chickens to a final virus titer of 107 EID50/ml. Then, we loaded 250 μl of the prepared sample into a CNT-STEM of 95-nm intertubular distance and extracted RNA into a final volume of 50 μl for NGS analysis. Compared with control RNA extracted from 50 μl of original H5N2 sample, both the concentrations of the total RNA and the converted complementary DNA (cDNA) were higher after the CNT-STEM enrichment and concentration (RNA, 870 ± 50 pg/μl versus 144 ± 34 pg/μl; cDNA, 3.8 nM versus 0.8 nM). The NGS viral reads increased from 2.9% (37,627 reads) to 90.6% (1,175,537 reads), thus corresponding to an enrichment factor of ~600 and indicating that the CNT-STEM removed most of the contamination from the chicken host at the same time (Fig. 6A). For the CNT-STEM–processed sample, by following the bioinformatics pipeline in fig. S10, the viral reads by NGS were de novo assembled into eight single contiguous sequences (contigs) with an ~105× coverage. The nucleotide BLAST search to GenBank [nonredundant nucleotide (nr/nt) database] shows the assembled sequences form the complete genome of the unsequenced H5N2 LPAIV strain (Fig. 6B and table S2). High sequence coverage allowed us to identify 38 intrahost variants, including 35 intrahost single-nucleotide variation (iSNV) sites, 2 intrahost multiple-nucleotide variation sites, and 1 deletion site (data file S1). By searching through sequenced AIV strains in GenBank, the closest strain is H5N2 AIV strain A/mallard/Wisconsin/411/1981. Phylogenetic analysis of hemagglutinin (HA) and neuraminidase (NA) genes suggested that this H5N2 strain (A/chicken/PA/7659/1985) belongs to the same branch of H5N2 strains isolated during the 1980s in the eastern and midwestern United States (Fig. 6C and table S3). We named this unsequenced H5N2 strain A/chicken/PA/7659/1985 and deposited the sequence into the National Center for Biotechnology Information (NCBI) database under KP674444-KP674451 (eight segments, complete sequences). This H5N2 strain has the monobasic cleavage site (PQRETR/GLF) in the HA gene, indicating that it is an LPAIV, which can grow only in limited areas of the poultry host (49).
Fig. 6. CNT-STEM prepares mimic field samples for NGS virus analysis.
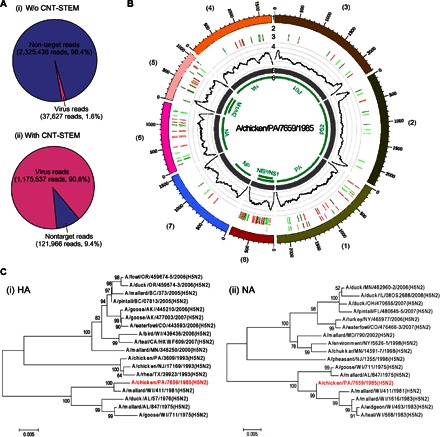
(A) Raw reads generated by NGS without and with CNT-STEM enrichment. (B) Circos plots of assembled contigs generated from NGS reads of the CNT-STEM–enriched H5N2 LPAIV samples. Track 1 (outermost), scale mark; track 2, iSNV; track 3, variants comparing to H5N2 AVI strain A/mallard/WI/411/1981; track 4, coverage (black) on scale of 0 to 30,000 reads; track 5, de novo assembled contigs after CNT-STEM enrichment (gray); track 6, open reading frames (green); color coding in tracks 2 and 3: deletion (black), transition (A-G, fluorescent green; G-A, dark green; C-T, dark red; T-C, light red), transversion (A-C, brown; C-A, purple; A-T, dark blue; T-A, fluorescent blue; G-T, dark orange; T-G, violet; C-G, yellow; G-C, light violet). (C) Phylogenetic tree plots generated by comparing the HA (i) and NA (ii) genes of the sequenced H5N2 AIV (highlighted in red) to those of closely related AIV strains isolated in North America from GenBank.
Field sample validation—A case study of AIV surveillance
To validate our new approach for real field samples, we applied a cloacal swab pool collected from five ducks during the 2012 AIV surveillance in Pennsylvania. The sample was previously detected as AIV type A–positive by rRT-PCR. Without any virus purification and propagation, 1.0 ml of the total ~5-ml suspension of the duck swab sample was enriched and concentrated by a CNT-STEM of 95-nm intertubular distance. No clogging was observed under SEM (fig. S11). Measured by rRT-PCR, the CNT-STEM increased virus titer from 6 × 102 to 2 × 104 EID50/ml (fig. S12). NGS and de novo sequence assembly yielded eight AIV contigs in complete lengths (Fig. 7A), but no AIV-related contig was discovered in the sample without CNT-STEM enrichment. The nucleotide BLAST search of GenBank (nr/nt database) showed that the sequenced AIV was an unsequenced strain and had different homologies to other reported strains, with ~99% similarities to the closest strains (table S4). Phylogenetic analysis indicated that the sample is an emerging H11N9 strain. It is closest to two H11N9 strains, A/duck/MN/Sg-00118/2007 (H11N9) and A/pintail/MN/Sg-00149/2007 (H11N9), isolated in Minnesota, USA, in 2007 (Fig. 7B, tables S4 and S5, and data file S1). We named it “A/duck/PA/02099/2012 (H11N9),” and the sequence was deposited in the NCBI database under KR870234-KR870241 (eight segments, complete sequences). The H11N9 strain was further confirmed by the U.S. Department of Agriculture–National Veterinary Services Laboratories (Ames, IA) through serologic tests.
Fig. 7. Identification of an emerging AIV H11N9 strain from a surveillance swab sample using CNT-STEM followed by NGS and de novo genome sequence assembly.
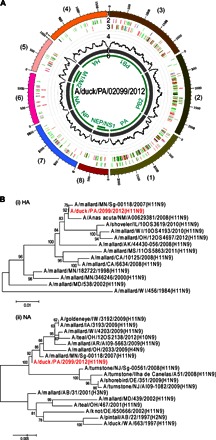
(A) Circos plot of assembled H11N9 contigs generated by NGS from a CNT-STEM–enriched wild duck swab pool. Track 1 (outermost), scale mark; track 2, identified iSNV; track 3, variants compared to a previously reported H11N9 AIV strain (A/duck/MN/Sg-00118/2007); track 4, coverage on scale of 0 to 50 reads (black); track 5, de novo assembled contigs after CNT-STEM enrichment (gray); track 6, open reading frames (green); color coding in tracks 2 and 3: deletion (black), transition (A-G, fluorescent green; G-A, dark green; C-T, dark red; T-C, light red), transversion (A-C, brown; C-A, purple; A-T, dark blue; T-A, fluorescent blue; G-T, dark orange; T-G, violet; C-G, yellow; G-C, light violet). (B) Phylogenetic tree plots generated by comparing the HA and NA genes of the sequenced H11N9 AIV (highlighted in red) to selected closely related AIV strains isolated in North America from GenBank.
Field sample validation—A case study of an unknown turkey virus
To verify the utility of our novel method with a truly clinically “unknown” virus, we used the CNT-STEM to process an eyelid tissue homogenate from a clinical case of a turkey reported to the Penn State Animal Diagnostic Laboratory in the summer of 2014. The turkeys had a symptom of blepharoconjunctivitis that had nodules and swollen lesions and was suspected to be caused by a viral agent. Various common tests for virus identification based on the symptoms of the infected turkeys, such as general serologic tests (for example, fluorescent antibody, agar gel immunodiffusion, hemagglutination inhibition, and virus neutralization) and molecular assays (for example, PCR), came out negative. We used CNT-STEM with NGS as the last resort. First, 5 ml of tissue homogenate was filtered through a membrane filter of 0.45-μm pore size. Then, 750 μl of filtrate was enriched and concentrated to 50 μl by a CNT-STEM of 25-nm intertubular distance and was analyzed by NGS. From the CNT-STEM–processed sample, 3.81% of the total NGS reads were viral reads (50,076 of 1,263,289), in contrast to only 0.001% of viral reads (17 of 1,626,134) from 50 μl of the original membrane filter tissue filtrate without CNT-STEM enrichment/concentration. The NGS reads represent an enrichment factor of 3.8 × 103. After assembly, two viral contigs were obtained with an average coverage of 1056. The nucleotide BLAST search identified this putative viral agent as a new variant strain of infectious bursal disease virus (IBDV) with less than 94 to 95% similarity to other reported IBDV strains in the United States (Fig. 8A). iSNV analysis identified four iSNV sites, where two iSNVs resulted in amino acid changes. Phylogenetic analysis and BLAST search results indicated that this is a novel strain (IBDV/turkey/PA/00924/14), close to an IBDV strain of serotype 2 isolated from turkeys in Ohio, USA (Fig. 8B, tables S6 and S7, and data file S1). Serotype 2 is relatively rare for sequenced and identified IBDVs, and we suspect that this is the reason why initial serologic and molecular tests failed to identify the virus. Moreover, the viral agent was observed under TEM, and it consisted of “birnavirus-like” particles of ~65 nm in diameter, well matched with the IBDV identification (Fig. 8B, inset). We name the virus “IBDV/turkey/PA/00924/14” and deposited the sequence with NCBI database under KP642112 (segment A) and KP642111 (segment B).
Fig. 8. Identification of a new IBDV strain from a turkey eyelid tissue sample using CNT-STEM followed by NGS and de novo genome sequence assembly.
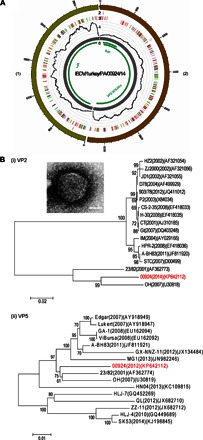
(A) Circos plots of assembled contigs generated from NGS reads of the CNT-STEM–enriched IBDV samples. Track 1 (outermost), scale mark; track 2, iSNV; track 3, variants comparing to OH strain (U30818) (b); track 4, coverage (black) on scale of 0 to 2625 reads; track 5, de novo assembled contigs after CNT-STEM enrichment (gray); track 6, open reading frames (green); color coding in tracks 2 and 3: deletion (black), transition (A-G, fluorescent green; G-A, dark green; C-T, dark red; T-C, light red), and transversion (A-C, brown; C-A, purple; A-T, dark blue; T-A, fluorescent blue; G-T, dark orange; T-G, violet; C-G, yellow; G-C, light violet). (B) Phylogenetic tree plots generated by comparing the open reading frames VP2/VP3/VP4 (i) and VP1 (ii) of IBDV/turkey/PA/00924/14 (highlighted in red) to previously reported IBDVs.
DISCUSSION
CNTs are among the strongest materials on Earth (38, 50). The strength and stiffness of N-MWCNTs are comparable to pristine MWCNTs (51). Because filtration is mainly a mechanical process, the high stiffness of the constructing nanomaterial enables us to make a device with extremely high porosity up to 95% while still maintaining structure integrity during filtration. Therefore, the robustness of CNTs and the extremely high porosity of the N-MWCNT arrays distinguish our CNT-STEM technology from other existing ultrafiltration techniques; at least two orders of magnitude lower in normalized flow resistance compared with commercial ultrafiltration membranes (table S8). This high porosity is critical for reducing flow resistance, preventing device clogging, and decreasing CNT material usage to minimize negative effect in downstream virus analyses, all of which empower the CNT-STEM as a point-of-care platform for efficient virus sample preparation from animal and human samples.
It is also noteworthy that the overall success rate from device fabrication to testing is 76.8% of 228 fabricated devices for these studies (see note S2). Device failure is due to leakage (13.6%), misalignment of PDMS-CNTs (7.0%), and N-MWCNT structure inhomogeneities (2.6%) (fig. S14). However, all of these can be improved by further microfabrication tuning. For leakage, we developed a simple and effective method to evaluate it (before introducing real samples), by measuring the flow rate of buffer solution through the CNT-STEM device.
The tunable range of the intertubular distance of N-MWCNT (17 to 325 nm) spans most of the virus size spectrum and provides unique flexibility in device design/fabrication able to reach the best performance for different viruses. To prepare samples for NGS, it is preferable to use CNT-STEM with larger intertubular distance if host ribosome RNA (rRNA) is a concern; larger intertubular distance will not trap ribosomes (~20 nm in diameter). Thus, we used CNT-STEM with 95-nm intertubular distance for the AIV samples targeted for NGS analysis. This is also justified for mimicking H5N2 swab samples: the rRNA reads reduced from 985,397 (41.7% of total reads) to 33,735 (2.6% of total reads) after the CNT-STEM sample preparation. For viruses of small size or samples with unknown viruses, it is more preferable to test viruses with devices of smaller intertubular distance. We used CNT-STEM of 25-nm intertubular distance to enrich and concentrate unknown viruses from the turkey eyelid tissue sample, and it turned out that the isolated IBDV was smaller than the AIV we used to work with (65 nm versus 93 nm).
CNT-STEM does not require antibody or any predesigned probe to recognize the viruses. This constitutes a unique advantage for virus discovery, and diagnosis of emerging new viruses and clinical cases of rare virus infection, in which the corresponding antibody may be not available. The selectivity of the CNT-STEM is provided by designing the intertubular distance to match the size of the targeted virus. The results show that CNT-STEM has a capture efficiency of 96.5 ± 0.5% and 88.0 ± 0.3% (Fig. 4C), when we used CNT-STEM of 25- and 95-nm intertubular distance to capture AIV, respectively. The capture efficiency is better or comparable to highly efficient immune-based capture (52–57).
To verify whether macrobiomolecules (larger than 10 nm) can pass through CNT-STEM, we prepared fluorescein isothiocyanate (FITC)–conjugated immunoglobulin G (IgG; Abcam, no. ab20921) with a concentration of 1 μg/ml and applied to CNT-STEM with 25-nm intertubular distance (fig. S15). We measured the penetration ratio as 98 ± 5% (n = 4). The result indicates that macrobiomolecules, such as IgG with a size smaller than intertubular distance, can pass through CNT-STEM without being trapped.
It has been reported that a high concentration of CNTs can inhibit PCR, whereas a low concentration of CNTs may enhance it. Our experiments suggest that there was no noticeable effect of N-MWCNT on the cycle threshold (Ct) values of the rRT-PCR. We measured the weight of N-MWCNTs inside each CNT-STEM as 26 μg, which corresponds to a final concentration of 0.5 μg/μl in the rRT-PCR mixture. The concentration is consistent with the previously reported CNT concentration ranges that have no effect or can enhance PCR.
In both the rRT-PCR virus detection and the ECE virus isolation experiments, the improvement correlates with the volume ratio of the original sample to that of the resuspended sample after enrichment, which underlines the importance of the optimal sample concentration provided by the CNT-STEM. Concentration effects can account for a large part of the improvement of rRT-PCR and virus isolation, because the contaminating materials do not significantly affect the highly specific rRT-PCR virus detection and they are nonproliferable in ECEs. However, the contaminant removal and sample concentration are key for the whole-genome sequencing using NGS, because random primers that do not distinguish viral targets from other contaminating genetic materials are used.
The CNT-STEM reported here provides a unique platform of a nanomaterial-integrated microfluidic device for label-free enrichment and concentration of viruses from field samples. By engineering the bottom-up synthesis process, we selectively grew N-MWCNT arrays on device substrates and then integrated them directly into microfluidic devices. This combined bottom-up synthesis and top-down microfabrication makes the production of the device potentially scalable and low cost. The unique properties of the vertically aligned N-MWCNT enable the CNT-STEM to enrich viable virus particles and remove host and environmental contaminants in a highly efficient way. The tunable size range of the CNT-STEM covers the size of most of the reported viruses. We demonstrated that this novel technology can significantly improve current rRT-PCR and virus isolation in AIV surveillance. It enables genomic sequencing using NGS directly from real field samples without virus amplification. Because neither CNT-STEM–based virus sample preparation nor NGS requires previous knowledge of the viruses inside the sample, this combination provides a unique and powerful approach to novel and emerging virus discovery, thus significantly contributing to the control and eradication of viral infectious diseases.
In the future, we believe that the aligned CNTs used in our devices could be chemically functionalized with biomolecules (for example, biotin) (40) or other chemical groups (58–61). This could potentially improve selectivity of our CNT-STEMs, especially when processing multivirus samples with an overlapped size distribution. Another direction is to integrate a detection system with the CNT-STEM for real-time virus detection, such as on-chip rRT-PCR. An integrated CNT-STEM system could potentially provide a rapid pathogen detection and a real-time quantification.
MATERIALS AND METHODS
Patterning of iron catalyst thin film
As shown in fig. S1A, a 4-inch single-side polished prime silicon wafer was cleaned in piranha solution, acetone, isopropyl alcohol, and ultrapure water sequentially. Lift-off photoresist LOR5A (MicroChem) and negative photoresist NFR105G (JSR Microelectronics) were spin-coated at 4000 rpm, respectively, followed by photolithography patterning with a contact aligner (Karl Suss MA/BA6). Iron was deposited by an e-beam evaporator (Semicore) under 10−7 mtorr of vacuum with deposition rate of 0.1 nm per second to target thicknesses of 1, 3, 5, 8, and 10 nm. The actual thicknesses of the deposited films were measured to be 1.0 ± 0.1, 3.0 ± 0.2, 6.5 ± 0.5, 9.2 ± 0.4, and 11.9 ± 0.8 nm by atomic force microscopy (Bruker Dimension Icon). A thin layer of negative photoresist NFR105G was spin-coated as a protective layer before dicing. The silicon substrate was then diced into individual dies of 1.2 cm × 1.2 cm by a dicing saw (Advanced Dicing Technologies). Photoresist was lifted off by soaking the substrate inside remover PG (MicroChem) overnight at 60°C.
N-MWCNT synthesis by AACVD
The AACVD setup consisted of an ultrasonic nebulizer (RBI-Instrumentation), two tube furnaces (Thermo Scientific) arranged in series, and a waste trap filled with acetone (fig. S2). Benzylamine (Fluka, CAS no. 100-46-9) was fed through the system by a nebulizer, working as both carbon source and nitrogen dopant. The iron-patterned substrates were placed in the second furnace. All components were airtight-sealed by silicone paste (McMaster-Carr) and flushed with argon and 15% hydrogen of flow rate (0.5 liter/min) for 5 min. The furnace temperature ramped to 825°C in 30 min. When the temperature reached 825°C, we turned on the nebulizer and increased the argon and 15% hydrogen flow to 2.5 liter/min. After the synthesis process was completed, we turned off the nebulizer, decreased the flow rate of carrier gas back to 0.5 liter/min, and set the furnace temperature back to 25°C. The cooling process usually took 3 hours for the furnaces to reach room temperature.
CNT-STEM assembly and experimental setup
The N-MWCNT forest pattern was designed as a droplet shape to evenly distribute the aqueous sample. The patterned N-MWCNT structure on the silicon substrate was integrated into a microfluidic device by bonding it with a PDMS chamber. The PDMS chamber was fabricated by standard soft lithography (62). The mold was fabricated by patterning SU-8 on silicon wafers with a contact aligner (Karl Suss MA/BA6). The ratio of the depth of the PDMS chamber and the height of the N-MWCNT forest was ~0.8 to 0.9. Then, well-mixed PDMS precursor (part A/part B, 10:1; Sylgard 184, Dow Corning) was poured onto the SU-8 mold and baked at 60°C for 40 min. The partially cured PDMS layer was diced into 1.2-cm × 1.2-cm squares with a razor blade. Two through-holes serving as inlet and outlet, 4 and 1 mm in diameter, respectively, were punctured through the PDMS using a luer adapter (BD). Before bonding, both the PDMS chamber and the N-MWCNT–patterned silicon substrate were treated with radio frequency oxygen plasma (M4L, PVA TePla Inc.) with processing parameters of 400-mtorr oxygen pressure, 50-W power, and 30-s duration. Then, they were aligned and gently pressed together and baked at 85°C for 4 hours.
The experimental system for the CNT-STEM included a sample reservoir, a waste trap, and components for flow actuation and control (fig. S1B). A 100-μl cylindrical sample reservoir formed with silicone tube of 5-mm inner diameter was attached to the inlet port. The outlet port was connected to a custom-made waste trap using a silicone tube of 0.5-mm inner diameter. The waste trap had three connections to a vacuum pump, a miniaturized pressure sensor, and a mechanical pressure regulator, respectively. The vacuum pump (McMaster-Carr) provided a negative pressure. The miniaturized pressure sensor (Honeywell ASDXL) had a sensing range of 10 inches of water column. It was soldered to a printed circuit board (McMaster-Carr) and powered by a 9-V battery. The mechanical pressure regulator (McMaster-Carr) controlled the vacuum suction. Fluidic connections were sealed by applying uncured PDMS precursor as sealant to the ends of the connections and then cured under room temperature for 24 hours.
Characterization of N-MWCNT forest geometries of vertically aligned N-MWCNT by AACVD synthesis
The nanoscale geometries of the N-MWCNT synthesized on 3-, 6.5-, and 12-nm-thick iron catalyst thin films under different synthesis times of 5, 10, 20, 30, and 40 min were studied by cross-sectional SEM images of the N-MWCNT structure taken by a field-emission SEM (FESEM) with accelerating voltage of 5 kV (LEO 1530 FESEM). The height of the N-MWCNT forest was characterized with cross-sectional SEM images under 8 × 102 magnification. For diameter analysis of single N-MWCNT, 6 × 104 magnification was used and a total number of 500 focused N-MWCNTs were measured with ImageJ. Both the N-MWCNT diameter D and its probability density function f(D) were calculated. For density and intertubular distance measurement of the aligned N-MWCNT structure, we took the images under 2.5 × 104 magnification at the bottom of the N-MWCNT forest close to the substrate. One line equivalent to 1 μm in length was drawn perpendicular to the N-MWCNT growth direction on each image. The numbers of focused N-MWCNTs that crossed the line were counted to calculate its linear density λ. Twenty images of each synthesis condition were analyzed, and five 1-μm lines were drawn for each image. For the intertubular distance, we measured the distance between pairs of neighboring focused N-MWCNTs that were crossed by the drawing line. Twenty images of each synthesis condition were collected, and data on five drawing lines on each image were analyzed. Assuming that the N-MWCNT density is isotropic in 2D, the porosity Φ can be calculated from the measured N-MWCNT line density λ, diameter Di, and the probability density function of the diameter f(Di): .
Measurement and analysis of the iron nanoparticle geometry
To study the geometrical properties of the iron nanoparticles and the relationship to those of the N-MWCNTs, we diced silicon wafers into device dies and deposited iron catalyst thin films of targeted thicknesses of 1, 3, 5, 8, and 10 nm on different device dies. For one set of device dies with different thicknesses of the iron catalyst film, the AACVD process went through the thermal ramping stage and was terminated before feeding the precursor (benzylamine). The SEM images of the top view of the iron nanoparticles were taken under 5 × 104 magnification. We analyzed SEM images of iron particles for their size and spatial distribution using MATLAB image processing toolbox. The average particle-to-particle distance was calculated by applying the Delaunay triangle selection to determine the closest neighbor particles, then represented as the mean of the three edges r1, r2, and r3 (fig. S16) (63).
Raman spectra measurement of N-MWCNT
AACVD-synthesized N-MWCNT was characterized by Raman microscopy (Renishaw, InVia Raman microscopy) using 514-nm laser excitation for 30 s under ×50 magnification. The laser power to the sample was 10 μW.
CNT-STEM filtration process
The assembled CNT-STEM was primed by adding 10 μl of 0.5% Tween 20 (Sigma-Aldrich) at the inlet port and letting the device sit undisturbed until all the air inside the device was replaced by Tween 20. This wetting process took around 15 min. Subsequently, another 50 μl of 0.5% Tween 20 was added to the inlet port. The vacuum suction from the outlet was turned on, and the differential pressure was maintained at 0.1 psi (6.9 × 102 Pa) to move the aqueous phase through the CNT-STEM. In the meantime, device leakage was tested by estimating the travel speed of the air-liquid interface inside the silicone tubing. If the device passed the leak test, then 200 μl of Dulbecco’s phosphate-buffered saline (DPBS) (Cellgro) was added to wash the device. After most of the DPBS flew through the device, we then added the virus sample to the inlet port while the vacuum suction remained. After most of the virus sample was filtered through, 50 μl of DPBS was added to rinse the device. All samples containing viruses were filtered through member filters (VWR) of 0.2- or 0.45-μm pore size for swab samples and tissue samples before introducing the filtrates using the CNT-STEMs.
N-MWCNT intertubular distance characterized by nanoparticle penetration
Fluorescein solution (Sigma-Aldrich, no. 46955) and polystyrene nanosphere suspensions (Thermo Scientific Inc.) were diluted by 0.5% Tween 20 into final concentration of 0.01% (solid). After device priming, 20 μl of the suspension was loaded at the inlet port. The vacuum suction was turned off after all the suspension was transported into the device. The fluorescence image of the device was taken by an sCMOS camera (Hamamatsu ORCA-Flash4.0 V2) connected to a fluorescence microscope (Olympus IX71). The fluorescence intensity was calibrated and measured by ImageJ (64). The penetration ratio was defined by the ratio of fluorescence intensity outside the CNT droplet-shaped chamber (Iout) to that inside (Iin), both corrected with background fluorescence intensity (Ibg) without the fluorescent agents: .
Size measurement of nanospheres and AIV virions
The diameters of fluorescent polystyrene nanospheres were measured by a Nano ZS particle size analyzer (Malvern Zetasizer, Malvern Instruments Ltd.). The size distribution of the nanospheres was calculated by the accompanying software (Nanov510) using a refractive index of 1.59.
Similarly, 107 EID50/ml of AIV solution was diluted by 1000-fold with 20 mM phosphate buffer at pH 7.4. The suspension was then passed through membrane filters of 0.45-μm (Celltreat scientific products) and 0.2-μm (VWR) pore size sequentially and then analyzed on a Nano ZS particle size analyzer (Malvern Zetasizer, Malvern Instruments Ltd.). By assuming a refractive index of 1.48 (65), the Nanov510 software converted intensity data into diameter measurements.
H5N2 AIV propagation and sample preparation
H5N2 AIV was propagated in SPF ECEs via allantoic cavity route inoculation at 9 to 11 days of age. The inoculated eggs were placed in a 37°C egg incubator for 72 hours. Then, the eggs were removed from the incubator and chilled at 4°C for 4 hours. Each egg was cracked open at the top air sac. The shell peeled without breaking the air sac membrane. The allantoic fluid containing the virus was harvested using a 3-ml sterile syringe with a 25-gauge × 5/8-inch needle. The harvested allantoic fluid was clarified by centrifugation at 8000 rpm for 5 min. The virus titers were measured in EID50 by the Reed-Muench method (66). Briefly, the freshly propagated H5N2 AIV stock was prepared in 10-fold serial dilutions from 101 to 109. Each dilution was inoculated into five eggs (0.1 ml per egg). The inoculated eggs were incubated at 37°C for 72 hours. The eggs were candled daily to remove dead eggs to chill them at 4°C refrigerator. After 72 hours of incubation, allantoic fluid was harvested from each egg and spun down. The supernatant was collected and passed through a membrane filter of 0.2-μm pore size before use. The infection status of each egg was determined by Dot-ELISA. AIV H5N2 samples were produced experimentally by spiking a freshly propagated LPAIV H5N2 strain (A/chicken/PA/7659/1985) into tracheal swabs obtained from SPF chickens.
On-chip immunofluorescence assay for H5N2 AIV detection and fluorescence intensity measurement
After virus capture and PBS washing inside the CNT-STEM, monoclonal antibody of the H5 HA protein (100 μl of 1:1000 diluted work solution; Penn State Animal Diagnostic Laboratory) was added through the inlet, incubated at 37°C for 40 min, and washed with 1 ml of PBS. Then, goat anti-mouse IgG conjugated with FITC (100 μl of 1:500 work dilution; KPL) was added and incubated at 37°C for 40 min, followed by 1 ml of PBS wash. Fluorescence microscopy images were obtained by an sCMOS camera (Hamamatsu ORCA-Flash4.0 V2) connected to a fluorescence microscope (Olympus IX71). By measuring average intensity of an area of 100 μm × 100 μm across the N-MWCNT walls, the fluorescence signal was calculated by ImageJ (64).
Virus lysis and RNA extraction
After performing virus filtration by the CNT-STEM, the virus-trapped device was disassembled by peeling the PDMS chamber using a razor blade. Normally, the N-MWCNT structure remained on the PDMS surface. We then scraped the N-MWCNT structure from the PDMS chamber with a razor blade and placed it into a microcentrifuge tube containing 50 μl of lysis/binding solution (MagMAX, Life Technologies). The viral RNA was extracted with MagMAX-96 AI/ND Viral RNA Isolation Kit (Life Technologies, catalog no. AM 1835) following the manufacturer’s protocol.
Real-time reverse transcription polymerase chain reaction
Primers and a probe specific to influenza type A were used (12). The master solution of rRT-PCR was prepared as a 50-μl reaction mixture using a OneStep RT-PCR Kit (Qiagen, catalog no. 210212), containing 10 μl of template RNA, 25 μl of ribonuclease (RNase)–free water, 10 μl of 5× buffer, 2 μl of deoxynucleotide triphosphate (dNTP) mix (10 mM for each dNTP), 1 μl of enzyme mix, and 1 μl of each of the two primers. Amplification was performed with a real-time PCR system (7300, Applied Biosystems Inc.) using a reverse transcription step at 50°C for 30 min. The initial PCR activation step was set at 95°C for 15 min, then followed by 94°C for 30 s, 50°C for 30 s, and 72°C for 90 s of each cycle for 38 cycles, and lastly finished with a single cycle of 72°C for 5 min. The data were collected and processed by the manufacturer’s software (7300 V1.4.0, Applied Biosystems Inc.). The Ct value was determined by the 2ΔΔCt method. The efficiency of the PCR was determined by , where Slope is the slope of the PCR standard curve.
Virus isolation and characterization of the propagation by Dot-ELISA
N-MWCNTs were collected into a microcentrifuge tube containing 0.1 ml of PBS and inoculated into an ECE as described for virus propagation. After 72 hours of incubation at 37°C, allantoic fluid was harvested. The allantoic fluid infection status was determined by Dot-ELISA using AIV H5 monoclonal antibody. We applied 10 μl of test specimens to a strip of a nitrocellulose membrane (Thermo Scientific Inc.). After the membrane was air-dried, we added blocking solution (10 ml of nonfat milk diluted in 30 ml of water) and incubated at room temperature for 10 to 20 min. After blocking, the membrane strip was allowed to air dry. Then, we applied anti-H5 monoclonal antibody and incubated for 60 min at room temperature, followed by PBS wash for 1 to 2 min with two repeats, and allowed to air dry. Next, we added goat anti-mouse IgG FITC conjugate at 1:500 working dilution of the stock solution (0.1 mg/ml) and incubated for 60 min at room temperature. We repeated the PBS wash step. Finally, we applied BCIP/NBT (5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium; Sigma-Aldrich, no. B8503) and kept the membrane in the dark during color development.
Sample preparation for NGS
To prepare a sample for NGS, we first built the cDNA library from the total RNA extracted from the sample. TruSeq Stranded Total RNA Sample Prep Kit (Illumina, catalog no. RS-122-2201) was used to reverse-transcribe the extracted RNA sample (total RNA) into cDNA. We followed instructions from the manufacturer with the exception that the initial poly A enrichment step was skipped. Briefly, by applying elevated temperatures, total RNA was fragmented into shorter segments. Those shorter RNA fragments were first reverse-transcribed into cDNA strands with random primers. By adding DNA polymerase I and RNase H, the complementary cDNA strands were synthesized. The cDNA was ligated to an adaptor and amplified by PCR to generate cDNA library. The quality of the cDNA library was tested by Agilent Bioanalyzer system (Agilent Technologies). Library concentration was assessed by real-time PCR using the KAPA Library Quantification Kit Illumina Platforms (Kapa Biosystems). Finally, the prepared cDNA libraries were loaded into different lanes of the MiSeq sequencer using 150-nt single-read sequencing (Illumina) to generate raw NGS reads.
De novo assembly and analysis
The overall pipeline for sequence data analysis is summarized in fig. S10. Starting from raw NGS reads, the added sequencing adaptors were removed by SortMeRNA. Quality trimming by Trimmomatic (BWA-MEM) was used to remove matched contaminating sequences of the host (for example, chicken, turkey, and human) and bacteria, as well as rRNA. Unmatched sequence reads were assembled using de novo assembly software SPAdes (V.3.5.0) (67) with K-mer size setting of 85. After de novo assembly, LASTZ (68) and SAMtools (69) were used to identify and obtain the final viral consensus sequences. Coverage was analyzed by BWA-MEM.
Phylogenetic analysis
Phylogenetic tree was generated by MEGA (V.6.06) (70) using ClustalW alignment and maximum likelihood (71). The tree topologies were made by bootstrap analysis with absolute distances following 1000 bootstrap replicates (72).
Intrahost variant calling and analysis
iSNVs were identified using a haplotype-based variant detector (Freebayes) with setting of ploidy of 1 and error rate of 0.8% (Illumina MiSeq) (73). If the frequency of variant population was higher than 0.8%, then it was considered as an iSNV site. The genetic variants were annotated by SnpEff v4.1 (74). The NGS data were displayed by generating plots with Circos (v 0.67) (75).
Field sample collection and preparation
AIV field samples were collected by inserting Dacron swabs (Fisherbrand, catalog no. 14-959-97B) into cloaca of poultry objects. The swabs were transferred into a cryovial containing 10 ml of viral transport medium (UTM, catalog no. 331C), which was prepared by following World Health Organization guidance. Before testing, a cryovial containing swab was first shaken by a vortex mixer (IKA MS2 S9 Mini Shaker) and then centrifuged under 1500g for 30 min. The supernatant was collected and passed through a membrane filter of 0.2-μm pore size before use.
The turkey tissue sample was from a turkey eyelid with gross lesion of swelling. The tissue sample was minced with sterile scissors in a 20-ml sterile plastic container (VWR, catalog no. 14310-684) containing viral transport medium at 1:5 (w/v) dilution. The minced tissue was transferred to a sterile Stomacher bag and homogenized in a Stomacher blender (Model 80, Seward Ltd.) for 2 to 3 min. The tissue homogenate was centrifuged at 1500 rpm for 10 min. The supernatant was filtered through a 0.45-μm syringe filter into a polypropylene conical tube, ready for virus detection.
Supplementary Material
Acknowledgments
We thank C. Praul for providing services of performing NGS at the Huck Institutes of the Life Sciences at the Pennsylvania State University. Funding: This research project was supported by a seed grant from the U.S. National Center for Research Resources and the National Center for Advancing Translational Sciences through an NIH grant (UL1 TR000127) to S.-Y.Z., M.T., and H.L.; a U.S. NIH Director’s New Innovator Award (DP2CA174508) to S.-Y.Z.; a U.S. Air Force Office of Scientific Research Multidisciplinary University Research Initiative grant (FA9550-12-1-0035) to M.T.; and a grant from the Pennsylvania State University College of Science to Y.-T.Y., N.P.-L., S.-Y.Z., and M.T. Author contributions: M.T. and S.-Y.Z. conceived and supervised the whole project. H.L. cosupervised virus-related parts. I.A. cosupervised NGS data analysis parts. Y.-T.Y. discovered the size-tunable CNT synthesis, designed droplet-shaped structures, fabricated CNT-STEM, characterized device performances, developed bioinformatics pipeline, and performed phylogenetic analysis. Y.T. prepared virus sample, designed virus-related experiments, performed virus detections (real-time reverse transcription PCR, Dot-ELISA, and egg inoculation), developed bioinformatics pipeline, and performed phylogenetic analysis. A.S. performed bioinformatics analysis and developed bioinformatics pipeline. A.D. synthesized CNTs and maintained chemical vapor deposition system. N.P.-L. helped in CNT synthesis. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from Y.-T.Y. (yxy155@psu.edu)
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/10/e1601026/DC1
fig. S1. Fabrication process and the testing setup of the CNT-STEM.
fig. S2. AACVD for N-MWCNT synthesis.
fig. S3. Raman spectra of the newly synthesized N-MWCNT structures on silicon substrates and the effect of the synthesis time on the height, diameter, and density of the aligned N-MWCNT structure.
fig. S4. Characterization of size-based particle capture by CNT-STEM.
fig. S5. Laser diffraction measurement of the size distribution of the LP AIV H5N2 strain used in this study.
fig. S6. Standard curve for the rRT-PCR detection of H5N2 AIV (n = 4 each).
fig. S7. Capture efficiency measurement of CNT-STEM with 25-, 95-, and 325-nm intertubular distances when loading H5N2 AIV of 106 EID50/ml of titer into each device (n = 6).
fig. S8. rRT-PCR curves of H5N2 AIV samples of 10 and 102 EID50/ml of titers without enrichment and those of 0.1 and 1 EID50/ml of titers with CNT-STEM enrichment (n = 6).
fig. S9. The compatibility test of N-MWCNT to rRT-PCR.
fig. S10. Diagram of data processing pipeline for NGS.
fig. S11. SEM images of CNT-STEM after processing field sample containing AIV.
fig. S12. rRT-PCR detection of the H11N9 AIV duck swab with and without CNT-STEM enrichment.
fig. S13. Structural mechanics analysis of N-MWCNT forest.
fig. S14. Analysis device yield, reliability, and failure modes.
fig. S15. Fluorescent image of FITC-conjugated IgG pass through CNT-STEM of 25-nm in interturbular distance.
fig. S16. Calculated distance between the iron particles based on the Delaunay triangle selection algorithm.
table S1. Measurement of the intertubular distance of N-MWCNT forest and the corresponding critical particle sizes of CNT-STEM.
table S2. Assembled contigs of the LP H5N2 AIV sample enriched by CNT-STEM.
table S3. Phylogenetic analysis of the sequenced H5N2 strain (A/chicken/PA/7659/1985) to closely related H5N2 AIV strains isolated from United States/Canada in GenBank.
table S4. Assembled contigs of the H11N9 AIV field sample enriched by CNT-STEM.
table S5. Phylogenetic analysis of the emerging H11N9 strain (A/duck/PA/02099/2012) to previously reported and closely related AIV strains.
table S6. Comparison of contigs of the unknown virus (IBDV/turkey/PA/00924/14) generated by de novo assembly after CNT-STEM enrichment and NGS to the closest IBDV strains in GenBank.
table S7. Single-nucleotide polymorphism/variant analysis of the “unknown” virus (IBDV/turkey/PA/00924/14) to sequenced IBDV virus strains.
table S8. Comparison of CNT-STEM to several reported ultrafiltration devices.
table S9. Yield and reliability analysis of CNT-STEM fabrication, assembly, and testing.
note S1. Structure stiffness of N-MWCNT forest in the CNT-STEM.
note S2. Device reliability study.
data file S1 (Microsoft Excel format)
REFERENCES AND NOTES
- 1.Fauci A. S., Morens D. M., The perpetual challenge of infectious diseases. N. Engl. J. Med. 366, 454–461 (2012). [DOI] [PubMed] [Google Scholar]
- 2.U.S. Department of Agriculture, Update on the Highly-Pathogenic Avian Influenza Outbreak of 2014–2015.
- 3.Anthony S. J., Epstein J. H., Murray K. A., Navarrete-Macias I., Zambrana-Torrelio C. M., Solovyov A., Ojeda-Flores R., Arrigo N. C., Islam A., Ali Khan S., Hosseini P., Bogich T. L., Olival K. J., Sanchez-Leon M. D., Karesh W. B., Goldstein T., Luby S. P., Morse S. S., Mazet J. A. K., Daszak P., Lipkin W. I., A strategy to estimate unknown viral diversity in mammals. mBio 4, e00598–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woolhouse M., Scott F., Hudson Z., Howey R., Chase-Topping M., Human viruses: Discovery and emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 2864–2871 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiu C. Y., Viral pathogen discovery. Curr. Opin. Microbiol. 16, 468–478 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howard C. R., Fletcher N. F., Emerging virus diseases: Can we ever expect the unexpected?. Emerg. Microbes Infect. 1, e46 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woolhouse M. E. J., Gowtage-Sequeria S., Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 11, 1842–1847 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pennington H., Politics, media and microbiologists. Nat. Rev. Microbiol. 2, 259–262 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King D. A., Peckham C., Waage J. K., Brownlie J., Woolhouse M. E. J., Infectious diseases: Preparing for the future. Science 313, 1392–1393 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Yolken R. H., Enzyme-linked immunosorbent-assay (ELISA): A practical tool for rapid diagnosis of viruses and other infectious agents. Yale J. Biol. Med. 53, 85–92 (1980). [PMC free article] [PubMed] [Google Scholar]
- 11.Ellis J. S., Zambon M. C., Molecular diagnosis of influenza. Rev. Med. Virol. 12, 375–389 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Spackman E., Senne D. A., Myers T. J., Bulaga L. L., Garber L. P., Perdue M. L., Lohman K., Daum L. T., Suarez D. L., Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 40, 3256–3260 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisfeld A. J., Neumann G., Kawaoka Y., Influenza A virus isolation, culture and identification. Nat. Protoc. 9, 2663–2681 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood J. M., Robertson J. S., From lethal virus to life-saving vaccine: Developing inactivated vaccines for pandemic influenza. Nat. Rev. Microbiol. 2, 842–847 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Radford A. D., Chapman D., Dixon L., Chantrey J., Darby A. C., Hall N., Application of next-generation sequencing technologies in virology. J. Gen. Virol. 93, 1853–1868 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beerenwinkel N., Günthard H. F., Roth V., Metzner K. J., Challenges and opportunities in estimating viral genetic diversity from next-generation sequencing data. Front. Microbiol. 3, 329 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chin C. D., Linder V., Sia S. K., Commercialization of microfluidic point-of-care diagnostic devices. Lab Chip 12, 2118–2134 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Heider S., Metzner C., Quantitative real-time single particle analysis of virions. Virology 462–463, 199–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L., Deng X., Mee E. T., Collot-Teixeira S., Anderson R., Schepelmann S., Minor P. D., Delwart E., Comparing viral metagenomics methods using a highly multiplexed human viral pathogens reagent. J. Virol. Methods 213, 139–146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noda T., Native morphology of influenza virions. Front. Microbiol. 2, 269 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Reis R., Zydney A., Bioprocess membrane technology. J. Membr. Sci. 297, 16–50 (2007). [Google Scholar]
- 22.Yeh Y.-T., Nisic M., Yu X., Xia Y., Zheng S.-Y., Point-of-care microdevices for blood plasma analysis in viral infectious diseases. Ann. Biomed. Eng. 42, 2333–2343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bibby K., Metagenomic identification of viral pathogens. Trends Biotechnol. 31, 275–279 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Daly G. M., Bexfield N., Heaney J., Stubbs S., Mayer A. P., Palser A., Kellam P., Drou N., Caccamo M., Tiley L., Alexander G. J. M., Bernal W., Heeney J. L., A viral discovery methodology for clinical biopsy samples utilising massively parallel next generation sequencing. PLOS One 6, e28879 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall R. J., Wang J., Todd A. K., Bissielo A. B., Yen S., Strydom H., Moore N. E., Ren X., Huang Q. S., Carter P. E., Peacey M., Evaluation of rapid and simple techniques for the enrichment of viruses prior to metagenomic virus discovery. J. Virol. Methods 195, 194–204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosseel T., Ozhelvaci O., Freimanis G., Van Borm S., Evaluation of convenient pretreatment protocols for RNA virus metagenomics in serum and tissue samples. J. Virol. Methods 222, 72–80 (2015). [DOI] [PubMed] [Google Scholar]
- 27.DiLeo A. J., Allegrezza A. E. Jr, Builder S. E., High resolution removal of virus from protein solutions using a membrane of unique structure. Nat. Biotechnol. 10, 182–188 (1992). [DOI] [PubMed] [Google Scholar]
- 28.Balasubramanian K., Burghard M., Biosensors based on carbon nanotubes. Anal. Bioanal. Chem. 385, 452–468 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Liu Z., Tabakman S., Welsher K., Dai H. J., Carbon nanotubes in biology and medicine: In vitro and in vivo detection, imaging and drug delivery. Nano Res. 2, 85–120 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai D., Mataraza J. M., Qin Z.-H., Huang Z., Huang J., Chiles T. C., Carnahan D., Kempa K., Ren Z., Highly efficient molecular delivery into mammalian cells using carbon nanotube spearing. Nat. Methods 2, 449–454 (2005). [DOI] [PubMed] [Google Scholar]
- 31.de Heer W. A., Châtelain A., Ugarte D., A carbon nanotube field-emission electron source. Science 270, 1179–1180 (1995). [Google Scholar]
- 32.Lee H. J., Park J., Yoon O. J., Kim H. W., Lee D. Y., Kim D. H., Lee W. B., Lee N.-E., Bonventre J. V., Kim S. S., Amine-modified single-walled carbon nanotubes protect neurons from injury in a rat stroke model. Nat. Nanotechnol. 6, 121–125 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xue X., Yang J.-Y., He Y., Wang L.-R., Liu P., Yu L.-S., Bi G.-H., Zhu M.-M., Liu Y.-Y., Xiang R.-W., Yang X.-T., Fan X.-Y., Wang X.-M., Qi J., Zhang H.-J., Wei T., Cui W., Ge G.-L., Xi Z.-X., Wu C.-F., Liang X.-J., Aggregated single-walled carbon nanotubes attenuate the behavioural and neurochemical effects of methamphetamine in mice. Nat. Nanotechnol. 11, 613–620 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen G. D., Fachin F., Fernandez-Suarez M., Wardle B. L., Toner M., Nanoporous elements in microfluidics for multiscale manipulation of bioparticles. Small 7, 1061–1067 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reyes-Reyes M., Grobert N., Kamalakaran R., Seeger T., Golberg D., Rühle M., Bando Y., Terrones H., Terrones M., Efficient encapsulation of gaseous nitrogen inside carbon nanotubes with bamboo-like structure using aerosol thermolysis. Chem. Phys. Lett. 396, 167–173 (2004). [Google Scholar]
- 36.Villalpando-Paez F., Zamudio A., Elias A. L., Son H., Barros E. B., Chou S. G., Kim Y. A., Muramatsu H., Hayashi T., Kong J., Terrones H., Dresselhaus G., Endo M., Terrones M., Dresselhaus M. S., Synthesis and characterization of long strands of nitrogen-doped single-walled carbon nanotubes. Chem. Phys. Lett. 424, 345–352 (2006). [Google Scholar]
- 37.Sumpter B. G., Meunier V., Romo-Herrera J. M., Cruz-Silva E., Cullen D. A., Terrones H., Smith D. J., Terrones M., Nitrogen-mediated carbon nanotube growth: Diameter reduction, metallicity, bundle dispersability, and bamboo-like structure formation. ACS Nano 1, 369–375 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Yu M.-F., Lourie O., Dyer M. J., Moloni K., Kelly T. F., Ruoff R. S., Strength and breaking mechanism of multiwalled carbon nanotubes under tensile load. Science 287, 637–640 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Mihalchik A. L., Ding W., Porter D. W., McLoughlin C., Schwegler-Berry D., Sisler J. D., Stefaniak A. B., Snyder-Talkington B. N., Cruz-Silva R., Terrones M., Tsuruoka S., Endo M., Castranova V., Qian Y., Effects of nitrogen-doped multi-walled carbon nanotubes compared to pristine multi-walled carbon nanotubes on human small airway epithelial cells. Toxicology 333, 25–36 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.González V. J., Gracia-Espino E., Morelos-Gómez A., López-Urías F., Terrones H., Terrones M., Biotin molecules on nitrogen-doped carbon nanotubes enhance the uniform anchoring and formation of Ag nanoparticles. Carbon 88, 51–59 (2015). [Google Scholar]
- 41.deHeer W. A., Bacsa W. S., Châtelain A., Gerfin T., Humphrey-Baker R., Forro L., Ugarte D., Aligned carbon nanotube films: Production and optical and electronic properties. Science 268, 845–847 (1995). [DOI] [PubMed] [Google Scholar]
- 42.Mizuno K., Ishii J., Kishida H., Hayamizu Y., Yasuda S., Futaba D. N., Yumura M., Hata K., A black body absorber from vertically aligned single-walled carbon nanotubes. Proc. Natl. Acad. Sci. U.S.A. 106, 6044–6047 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z.-P., Ci L., Bur J. A., Lin S.-Y., Ajayan P. M., Experimental observation of an extremely dark material made by a low-density nanotube array. Nano Lett. 8, 446–451 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Gao R., Cao B., Hu Y., Feng Z., Wang D., Hu W., Chen J., Jie Z., Qiu H., Xu K., Xu X., Lu H., Zhu W., Gao Z., Xiang N., Shen Y., He Z., Gu Y., Zhang Z., Yang Y., Zhao X., Zhou L., Li X., Zou S., Zhang Y., Li X., Yang L., Guo J., Dong J., Li Q., Dong L., Zhu Y., Bai T., Wang S., Hao P., Yang W., Zhang Y., Han J., Yu H., Li D., Gao G. F., Wu G., Wang Y., Yuan Z., Shu Y., Human infection with a novel avian-origin influenza A (H7N9) virus. N. Engl. J. Med. 368, 1888–1897 (2013). [DOI] [PubMed] [Google Scholar]
- 45.World Health Organization (WHO), “The world health report 2007 - A safer future: Global public health security in the 21st century” (WHO Press, Geneva, 2007). [Google Scholar]
- 46.Yang Z.-Y., Wei C.-J., Kong W.-P., Wu L., Xu L., Smith D. F., Nabel G. J., Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science 317, 825–828 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu H., Xie Z., Liu J., Lin L., Studies on multiplex RT-PCR for detection of Avian Influenza Virus type A group and specific H5 and H7 subtypes. J. Vet. Sci. Med. 1, 5 (2013). [Google Scholar]
- 48.Acevedo A., Andino R., Library preparation for highly accurate population sequencing of RNA viruses. Nat. Protoc. 9, 1760–1769 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D. J. Alexander, I. Capua, G. Koch, in Avian Influenza (Blackwell Publishing Ltd., 2009), pp. 217–237. [Google Scholar]
- 50.Qian D., Wagner G. J., Liu W. K., Yu M.-F., Ruoff R. S., Mechanics of carbon nanotubes. Appl. Mech. Rev. 55, 495–533 (2002). [Google Scholar]
- 51.Ganesan Y., Peng C., Lu Y., Ci L., Srivastava A., Ajayan P. M., Lou J., Effect of nitrogen doping on the mechanical properties of carbon nanotubes. ACS Nano 4, 7637–7643 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Hung L.-Y., Huang T.-B., Tsai Y.-C., Yeh C.-S., Lei H.-Y., Lee G.-B., A microfluidic immunomagnetic bead-based system for the rapid detection of influenza infections: From purified virus particles to clinical specimens. Biomed. Microdevices 15, 539–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao W., Zhang W.-P., Zhang Z.-L., He R.-L., Lin Y., Xie M., Wang H.-Z., Pang D.-W., Robust and highly sensitive fluorescence approach for point-of-care virus detection based on immunomagnetic separation. Anal. Chem. 84, 2358–2365 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Sakudo A., Onodera T., Virus capture using anionic polymer-coated magnetic beads (review). Int. J. Mol. Med. 30, 3–7 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Lien K.-Y., Hung L.-Y., Huang T.-B., Tsai Y.-C., Lei H.-Y., Lee G.-B., Rapid detection of influenza A virus infection utilizing an immunomagnetic bead-based microfluidic system. Biosens. Bioelectron. 26, 3900–3907 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lien K.-Y., Lin J.-L., Liu C.-Y., Lei H.-Y., Lee G.-B., Purification and enrichment of virus samples utilizing magnetic beads on a microfluidic system. Lab Chip 7, 868–875 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Jothikumar N., Cliver D. O., Mariam T. W., Immunomagnetic capture PCR for rapid concentration and detection of hepatitis A virus from environmental samples. Appl. Environ. Microbiol. 64, 504–508 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meng L., Fu C., Lu Q., Advanced technology for functionalization of carbon nanotubes. Prog. Nat. Sci. 19, 801–810 (2009). [Google Scholar]
- 59.V. K. Thakur, M. K. Thakur, Chemical Functionalization of Carbon Nanomaterials: Chemistry and Applications (CRC Press, 2015). [Google Scholar]
- 60.Balasubramanian K., Burghard M., Chemically functionalized carbon nanotubes. Small 1, 180–192 (2005). [DOI] [PubMed] [Google Scholar]
- 61.Han H., Wu X., Wu S., Zhang Q., Lu W., Zhang H., Pan D., Fabrication of alizarin red S/multi-walled carbon nanotube nanocomposites and their application in hydrogen peroxide detection. J. Mater. Sci. 48, 3422–3427 (2013). [Google Scholar]
- 62.Xia Y., Whitesides G. M., Soft lithography. Annu. Rev. Mater. Sci. 28, 153–184 (1998). [Google Scholar]
- 63.Bray D. J., Gilmour S. G., Guild F. J., Taylor A. C., Quantifying nanoparticle dispersion by using the area disorder of Delaunay triangulation. J. R. Stat. Soc. Ser. C Appl. Stat. 61, 253–275 (2012). [Google Scholar]
- 64.P. Bankhead, Analyzing Fluorescence Microscopy Images with ImageJ (Queen’s Univ., 2014). [Google Scholar]
- 65.Wang S., Shan X., Patel U., Huang X., Lu J., Li J., Tao N., Label-free imaging, detection, and mass measurement of single viruses by surface plasmon resonance. Proc. Natl. Acad. Sci. U.S.A. 107, 16028–16032 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reed L. J., Muench H., A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 27, 493–497 (1938). [Google Scholar]
- 67.Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., Lesin V. M., Nikolenko S. I., Pham S., Prjibelski A. D., Pyshkin A. V., Sirotkin A. V., Vyahhi N., Tesler G., Alekseyev M. A., Pevzner P. A., SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.R. S. Harris, Improved Pairwise Alignment of Genomic DNA (ProQuest, 2007). [Google Scholar]
- 69.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumar S., Tamura K., Nei M., MEGA: Molecular evolutionary genetics analysis software for microcomputers. Comput. Appl. Biosci. 10, 189–191 (1994). [DOI] [PubMed] [Google Scholar]
- 71.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S., MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Felsenstein J., Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791 (1985). [DOI] [PubMed] [Google Scholar]
- 73.Quail M. A., Smith M., Coupland P., Otto T. D., Harris S. R., Connor T. R., Bertoni A., Swerdlow H. P., Gu Y., A tale of three next generation sequencing platforms: Comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13, 341 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., Wang L., Land S. J., Lu X., Ruden D. M., A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Krzywinski M., Schein J., Birol İ., Connors J., Gascoyne R., Horsman D., Jones S. J., Marra M. A., Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bohonak D. M., Zydney A. L., Compaction and permeability effects with virus filtration membranes. J. Membrane Sci. 254, 71–79 (2005). [Google Scholar]
- 77.Van Holten R. W., Ciavarella D., Oulundsen G., Harmon F., Riester S., Incorporation of an additional viral-clearance step into a human immunoglobulin manufacturing process. Vox Sang. 83, 227–233 (2002). [DOI] [PubMed] [Google Scholar]
- 78.Yang S. Y., Ryu I., Kim H. Y., Kim J. K., Jang S. K., Russell T. P., Nanoporous membranes with ultrahigh selectivity and flux for the filtration of viruses. Adv. Mater. 18, 709–712 (2006). [Google Scholar]
- 79.Desai T. A., Hansford D. J., Leoni L., Essenpreis M., Ferrari M., Nanoporous anti-fouling silicon membranes for biosensor applications. Biosens. Bioelectron. 15, 453–462 (2000). [DOI] [PubMed] [Google Scholar]
- 80.Mueller J. A., Culley A. I., Steward G. F., Variables influencing extraction of nucleic acids from microbial plankton (viruses, bacteria, and protists) collected on nanoporous aluminum oxide filters. Appl. Environ. Microbiol. 80, 3930–3942 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Palacio L., Prádanos P., Calvo J. I., Hernández A., Porosity measurements by a gas penetration method and other techniques applied to membrane characterization. Thin Solid Films 348, 22–29 (1999). [Google Scholar]
- 82.Thormann A., Teuscher N., Pfannmöller M., Rothe U., Heilmann A., Nanoporous aluminum oxide membranes for filtration and biofunctionalization. Small 3, 1032–1040 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Urase T., Yamamoto K., Ohgaki S., Effect of pore structure of membranes and module configuration on virus retention. J. Membrane Sci. 115, 21–29 (1996). [Google Scholar]
- 84.Syedain Z. H., Bohonak D. M., Zydney A. L., Protein fouling of virus filtration membranes: Effects of membrane orientation and operating conditions. Biotechnol. Prog. 22, 1163–1169 (2006). [DOI] [PubMed] [Google Scholar]
- 85.J. M. Gere, B. J. Goodno, Mechanics of Materials (Cengage Learning, ed. 8, 2012). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/10/e1601026/DC1
fig. S1. Fabrication process and the testing setup of the CNT-STEM.
fig. S2. AACVD for N-MWCNT synthesis.
fig. S3. Raman spectra of the newly synthesized N-MWCNT structures on silicon substrates and the effect of the synthesis time on the height, diameter, and density of the aligned N-MWCNT structure.
fig. S4. Characterization of size-based particle capture by CNT-STEM.
fig. S5. Laser diffraction measurement of the size distribution of the LP AIV H5N2 strain used in this study.
fig. S6. Standard curve for the rRT-PCR detection of H5N2 AIV (n = 4 each).
fig. S7. Capture efficiency measurement of CNT-STEM with 25-, 95-, and 325-nm intertubular distances when loading H5N2 AIV of 106 EID50/ml of titer into each device (n = 6).
fig. S8. rRT-PCR curves of H5N2 AIV samples of 10 and 102 EID50/ml of titers without enrichment and those of 0.1 and 1 EID50/ml of titers with CNT-STEM enrichment (n = 6).
fig. S9. The compatibility test of N-MWCNT to rRT-PCR.
fig. S10. Diagram of data processing pipeline for NGS.
fig. S11. SEM images of CNT-STEM after processing field sample containing AIV.
fig. S12. rRT-PCR detection of the H11N9 AIV duck swab with and without CNT-STEM enrichment.
fig. S13. Structural mechanics analysis of N-MWCNT forest.
fig. S14. Analysis device yield, reliability, and failure modes.
fig. S15. Fluorescent image of FITC-conjugated IgG pass through CNT-STEM of 25-nm in interturbular distance.
fig. S16. Calculated distance between the iron particles based on the Delaunay triangle selection algorithm.
table S1. Measurement of the intertubular distance of N-MWCNT forest and the corresponding critical particle sizes of CNT-STEM.
table S2. Assembled contigs of the LP H5N2 AIV sample enriched by CNT-STEM.
table S3. Phylogenetic analysis of the sequenced H5N2 strain (A/chicken/PA/7659/1985) to closely related H5N2 AIV strains isolated from United States/Canada in GenBank.
table S4. Assembled contigs of the H11N9 AIV field sample enriched by CNT-STEM.
table S5. Phylogenetic analysis of the emerging H11N9 strain (A/duck/PA/02099/2012) to previously reported and closely related AIV strains.
table S6. Comparison of contigs of the unknown virus (IBDV/turkey/PA/00924/14) generated by de novo assembly after CNT-STEM enrichment and NGS to the closest IBDV strains in GenBank.
table S7. Single-nucleotide polymorphism/variant analysis of the “unknown” virus (IBDV/turkey/PA/00924/14) to sequenced IBDV virus strains.
table S8. Comparison of CNT-STEM to several reported ultrafiltration devices.
table S9. Yield and reliability analysis of CNT-STEM fabrication, assembly, and testing.
note S1. Structure stiffness of N-MWCNT forest in the CNT-STEM.
note S2. Device reliability study.
data file S1 (Microsoft Excel format)


