Fig. 6. CNT-STEM prepares mimic field samples for NGS virus analysis.
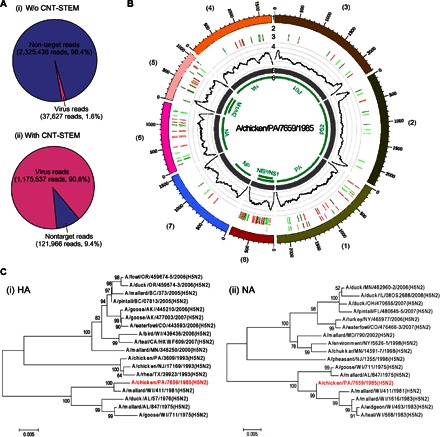
(A) Raw reads generated by NGS without and with CNT-STEM enrichment. (B) Circos plots of assembled contigs generated from NGS reads of the CNT-STEM–enriched H5N2 LPAIV samples. Track 1 (outermost), scale mark; track 2, iSNV; track 3, variants comparing to H5N2 AVI strain A/mallard/WI/411/1981; track 4, coverage (black) on scale of 0 to 30,000 reads; track 5, de novo assembled contigs after CNT-STEM enrichment (gray); track 6, open reading frames (green); color coding in tracks 2 and 3: deletion (black), transition (A-G, fluorescent green; G-A, dark green; C-T, dark red; T-C, light red), transversion (A-C, brown; C-A, purple; A-T, dark blue; T-A, fluorescent blue; G-T, dark orange; T-G, violet; C-G, yellow; G-C, light violet). (C) Phylogenetic tree plots generated by comparing the HA (i) and NA (ii) genes of the sequenced H5N2 AIV (highlighted in red) to those of closely related AIV strains isolated in North America from GenBank.
