Fig. 7. Identification of an emerging AIV H11N9 strain from a surveillance swab sample using CNT-STEM followed by NGS and de novo genome sequence assembly.
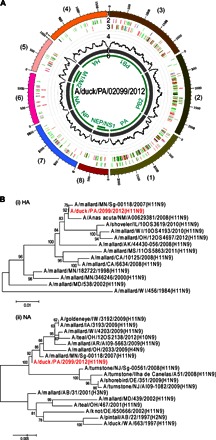
(A) Circos plot of assembled H11N9 contigs generated by NGS from a CNT-STEM–enriched wild duck swab pool. Track 1 (outermost), scale mark; track 2, identified iSNV; track 3, variants compared to a previously reported H11N9 AIV strain (A/duck/MN/Sg-00118/2007); track 4, coverage on scale of 0 to 50 reads (black); track 5, de novo assembled contigs after CNT-STEM enrichment (gray); track 6, open reading frames (green); color coding in tracks 2 and 3: deletion (black), transition (A-G, fluorescent green; G-A, dark green; C-T, dark red; T-C, light red), transversion (A-C, brown; C-A, purple; A-T, dark blue; T-A, fluorescent blue; G-T, dark orange; T-G, violet; C-G, yellow; G-C, light violet). (B) Phylogenetic tree plots generated by comparing the HA and NA genes of the sequenced H11N9 AIV (highlighted in red) to selected closely related AIV strains isolated in North America from GenBank.
