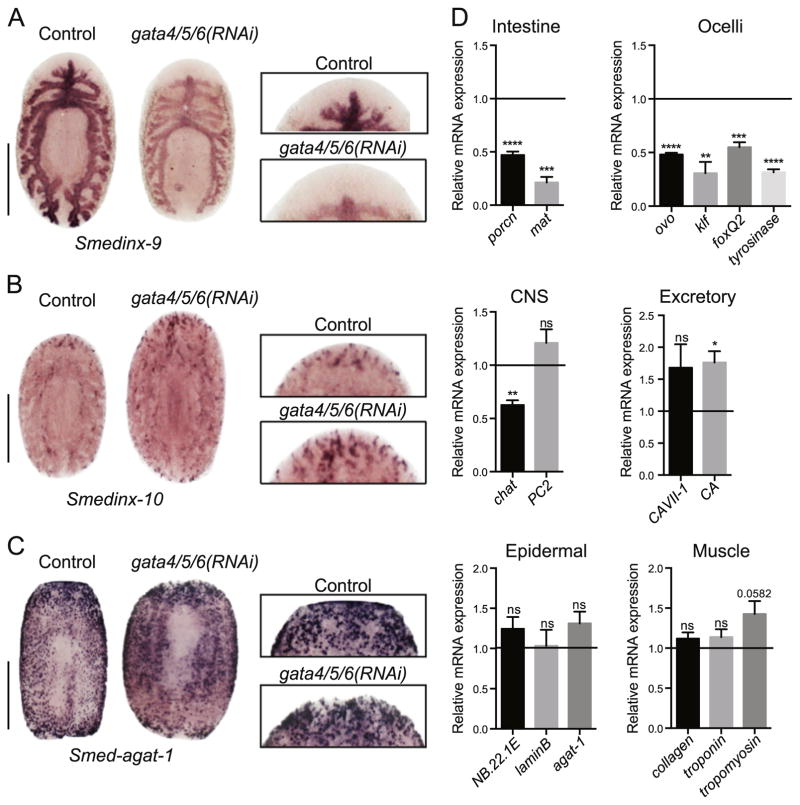
Fig. 3.
Analysis of differentiated cell lineages in Smed-gata4/5/6(RNAi) animals. (A) Representative WISH for smedinx-9, an intestine marker, in control and Smed-gata4/5/6 (RNAi) animals (anti-sense probe). A magnification of the blastema (inset) shows the absence of intestinal branching in the newly formed tissue in knock-down worms 7 days post-amputation. (n = 5) Scale bar: 250 μm. (B) Representative WISH for smedinx-10, an excretory marker (flame cells), in control and Smed-gata4/5/6(RNAi) animals (anti-sense probe). A magnification of the blastema (inset) shows that new flame cells are established in the newly-formed tissue 7 days post-amputation. (n = 5) Scale bar: 250 μm. (C) Representative WISH for Smed-agat-1, a general differentiation marker, in control and Smed-gata4/5/6(RNAi) animals (anti-sense probe). A magnification of the blastema (inset) shows that differentiated cell types are found in the new tissue 7 days post-amputation. (n = 5) Scale bar: 500 μm. (D) RT-qPCR analysis showing mRNA expression changes in differentiation tissue marker transcripts following Smed-gata4/5/6(RNAi) 7 days post-amputation. Bar graphs show fold change in expression relative to controls after normalization to GAPDH. Controls represented by horizontal line set at 1. Two-tailed unpaired Student’s t-test (****: p-value < 0.0001, ***: p-value < 0.001, **: p-value > 0.01*: p-value > 0.05, ns: not significant) values represent average and error bars s.e.m. Analysis for regenerating trunks based on 3 biological replicates (n = 15), each replicate containing 5 animals pooled.