Figure 1.
Single-Cell Transcriptomic Analysis of MERVL-LTR Driven Network Dynamics
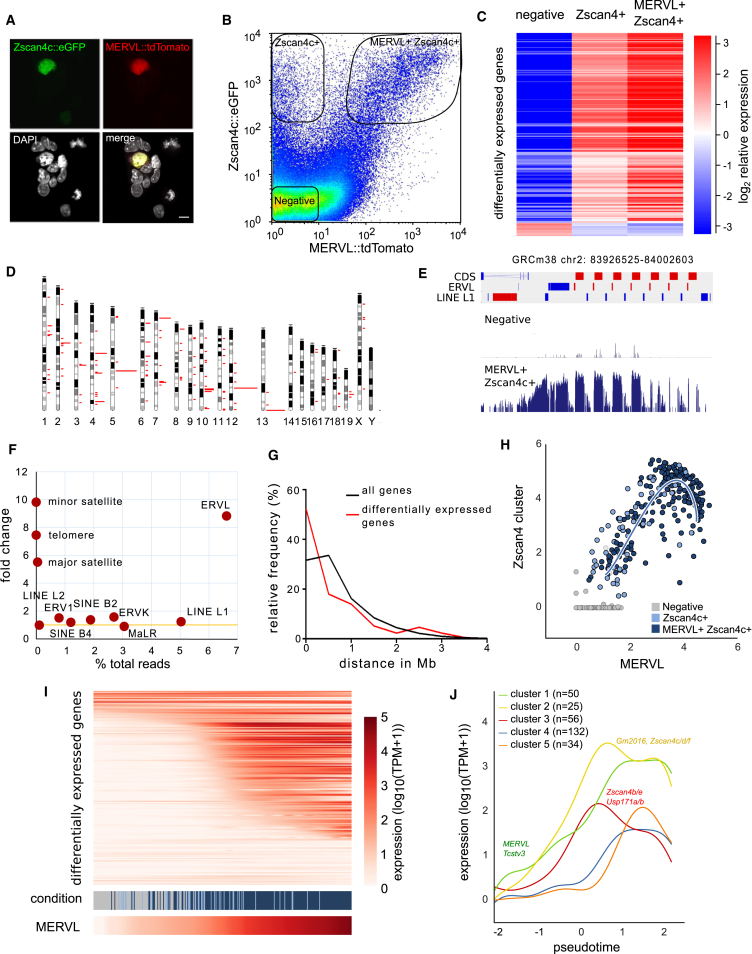
(A) Representative single z-section through a ESC colony containing a cell co-expressing the Zscan4c::EGFP (top left) and MERVL::tdTomato (top right) reporters. DNA is visualized with DAPI (bottom left). Scale bar, 10 μm.
(B) Flow cytometry plot of the MERVL::tdTomato (x axis) Zscan4c::eGFP (y axis) dual-reporter ESC line. Representative gates used to sort the three populations (negative, Zscan4c+, and Zscan4c+MERVL+) are shown.
(C) Heat map showing relative expression levels of 172 differentially expressed genes in negative (left column), Zscan4+ (middle column) and Zscan4+MERVL+ (right column) sorted populations as determined by total RNA-sequencing (see Supplemental Experimental Procedures). Scale bar depicts log2 relative expression.
(D) Overview showing localization and frequency of differentially expressed genes (red bars) across the genome.
(E) Schematic overview over the Gm13691 containing cluster on chromosome 2 depicting the organization of coding sequence (CDS), MERVL, and LINE L1 elements. Two data tracks show wiggle plots of total RNA-seq reads for MERVL+Zscan4+ (dark blue) and negative-sorted (gray) cells.
(F) Quantification of total RNA-seq reads mapping to different repeat classes in MERVL+Zscan4c+ and negative-sorted cells, plotted as fold change over percentage of total reads. The yellow line indicates no change.
(G) Relative frequency plot showing distance between gene and nearest MERVL promoter (MT2_Mm) of all genes (black) and differentially expressed genes (red). p value < 0.0001 Mann-Whitney U test.
(H) Log10 TPM (transcripts per million) values of MERVL (x axis) and the Zscan4 cluster (y axis) of single cells sorted from negative (gray), Zscan4c+ (light blue) and Zscan4c+MERVL+ (dark blue) gates. The solid blue line represents the projected trajectory of the cells in this two-dimensional space, or “pseudotime’ (see Supplemental Experimental Procedures).
(I) Smoothed heatmap showing expression of 172 differentially expressed genes (rows) across sorted single cells (columns) ordered by MERVL expression (bottom scale bar). Median Spearman rank correlation was 0.6 between MERVL and differentially expressed genes and −0.08 between MERVL and all genes.
(J) Expression profiles of dynamic clusters of genes across pseudotime, denoting selected genes of interest.