Figure 4.
Acute DNA Demethylation Is Not Sufficient for MERVL Network Activation
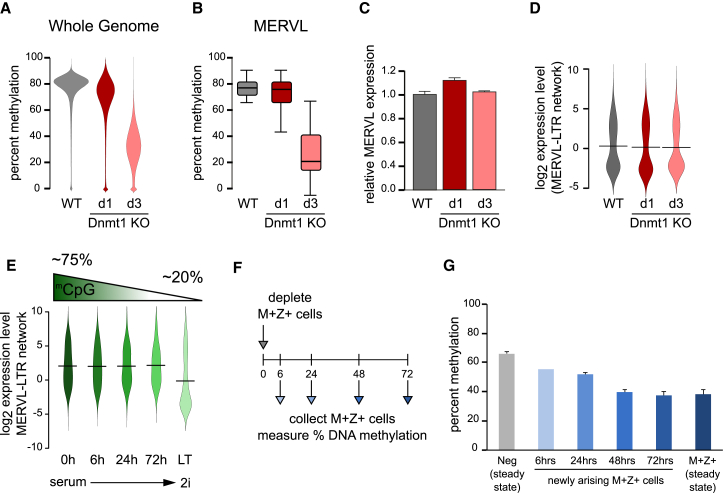
(A and B) Genome-wide (A) and MERVL (B) CpG methylation levels measured by bisulfite sequencing of wild-type (gray) and conditional Dnmt1 knockout ESCs induced for 1 day (dark red) or 3 days (pink).
(C) qRT-PCR analysis of MERVL expression in wild-type (gray) and conditional Dnmt1 knockout ESCs induced for 1 day (dark red) or 3 days (pink). Bars represent mean + SD of three biological replicates.
(D) Bean plots showing log2 expression levels of the MERVL-LTR-driven transcriptional network in wild-type (gray) and conditional Dnmt1 knockout ESCs induced for 1 day (dark red) or 3 days (pink).
(E) Bean plots showing log2 expression levels of the MERVL-LTR-driven transcriptional network in serum-treated (0 hr, dark green), and cells cultured in naive 2i conditions for 6 hr, 24 hr, 72 hr or long-term (LT; light green). Data were reanalyzed from von Meyenn et al. (2016). Average methylation levels are depicted above.
(F) Schematic showing release experiment in which MERVL+Zscan4c+ (M+Z+) cells are depleted from the steady-state population by flow cytometry sorting of the negative gate, followed by re-culturing. The “new” MERVL+Zscan4c+ (M+Z+) cells were subsequently collected by flow cytometry at the indicated time points for methylation analysis.
(G) Genome-wide methylation levels determined by PBAT analysis. Steady-state negative-sorted (light gray, first column) and MERVL+Zscan4c+ (M+Z+) sorted (dark blue, last column) cells are shown for reference. The other columns (light to dark blue) reflect cells that were initially sorted as negative, returned to culture for 6, 24, 48, or 72 hr, and then re-sorted as MERVL+Zscan4c+ (M+Z+) positive. Bars represent average of three biological replicates of 100 cells each ± SD, except the 6-hr time point (n = 1).
See also Figure S4.