Figure 2.
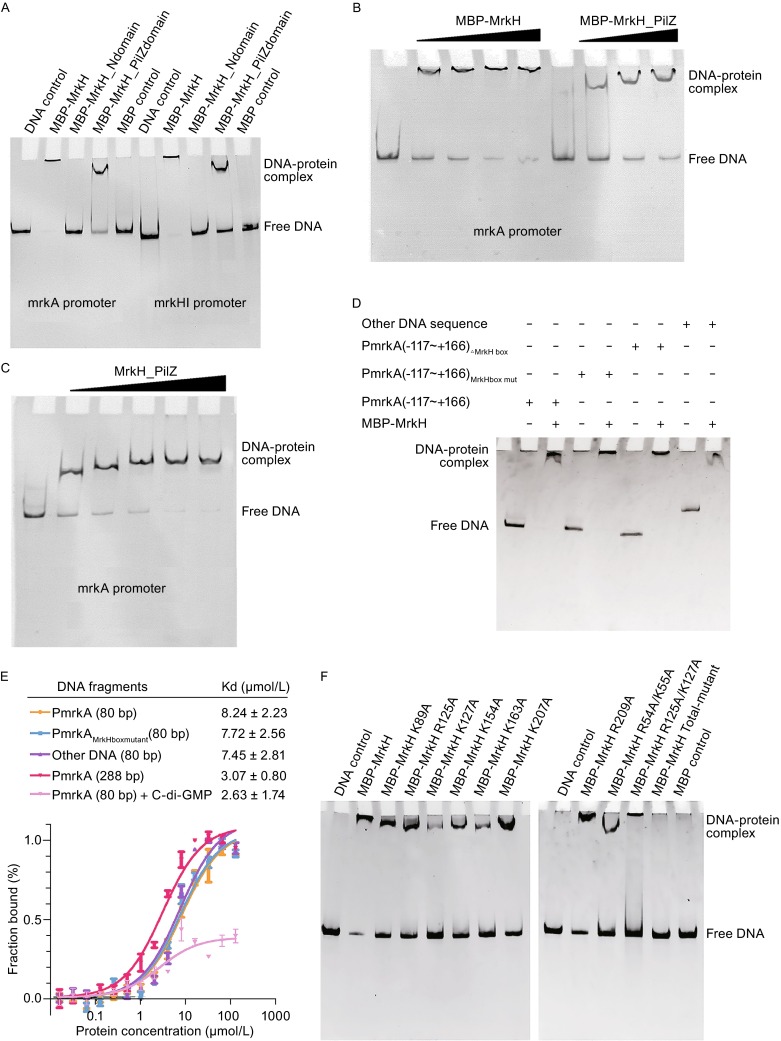
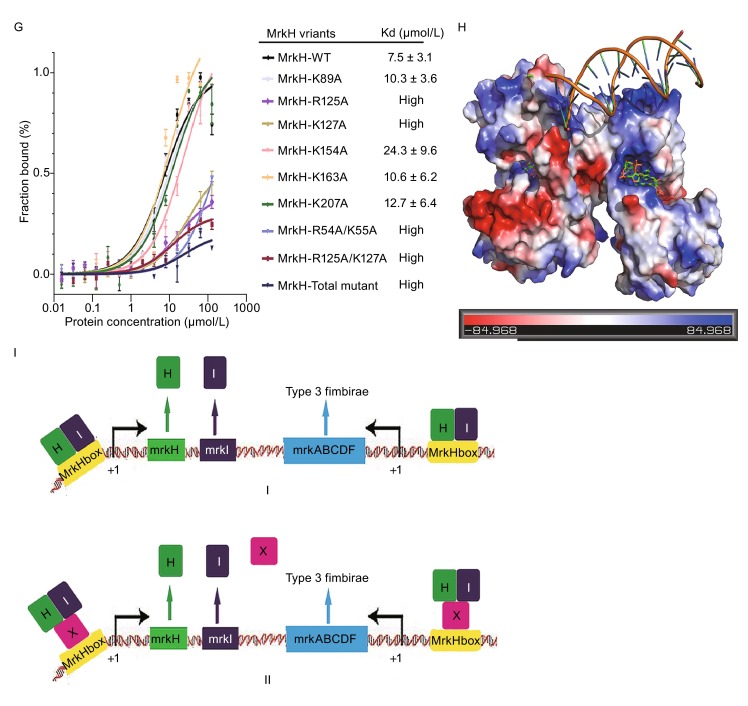
MrkH-PilZ domain is a novel DNA-binding motif. (A) EMSA was performed using a 288 bp mrkA promoter containing base pairs −117 to +166 relative to the start site of mrkA transcription and 250 bp mrkHI promoter containing base pairs −184 to +52 relative to the start site of mrkH transcription with MrkH and MrkH two sides domains. (B) EMSA was performed for MBP-MrkH and MBP-MrkH-PilZ with the 288 bp mrkA promoter sequence. The same amount of DNA was used for each lane. Free DNA decreases with the increase of protein concentration. (C) EMSA of MrkH-PilZ with the 288-bp mrkA promoter sequence. As protein concentration increases, the amount of free DNA drops,the electrophoretic mobility of protein-DNA complex also declines. (D) EMSA was performed for MBP-MrkH with a variety of 288 bp mrkA promoter sequences and a random DNA fragment. (E) Fluorescence polarization curves are shown for the binding of MrkH to the FAM-labeled mrkA promoter fragments (288 bps from −117 to +166; 80 bps from −117 to −37 and the ‘MrkH box’ mutant; and a random 80 bps DNA). (F) EMSA analysis of MBP-MrkH mutants and the mrkA promoter sequence. (G) DNA-binding affinities of wild-type and mutant MrkH. Fluorescence polarization curves are shown for the binding of purified proteins to a FAM-labeled 80 bp mrkA promoter fragment (from −117 to −37). (H) The Docking DNA/MrkH complex structure, in the proposed MrkH dimer two separate DNA-binding regions merge into a larger positively charged area binding DNA. (I) The model for MrkHI regulating the expression of type 3 fimbriae in K. pneumoniae.: [I]MrkH and MrkI form MrkHI complex which regulates the expression of mrkABCDF and mrkHI. [II] MrkHI complex recruits a not-yet-indentified protein X forming a ternary complex which regulates the transcription of mrkABCDF and mrkHI. The corresponding K d values are obtained by fitting data to one-site specific binding model, and the error bars represent ± SD for triplicate experiments
