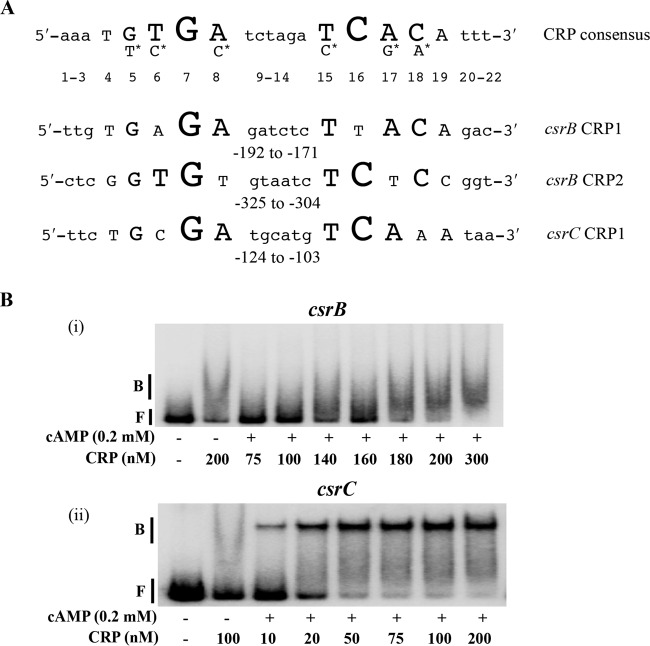
FIG 5.
Binding of cAMP-CRP to csrB and csrC DNA. (A) Consensus sequence for cAMP-CRP binding and predicted binding sites in the upstream flanking regions of csrB and csrC. Nucleotides in uppercase letters in the crp consensus sequence denote the conserved residues, and the size of the nucleotide letters indicates the relative degree of conservation of the nucleotide residue (47). Nucleotides with the asterisk indicate the second most highly conserved residue in comparison to the nucleotide under which they are placed. Numbers indicate the position of each nucleotide relative to the 5′ end of the consensus. Numbers beneath the csrB and csrC sequences are indicative of location with respect to the transcription initiation site. (B) Electrophoretic gel mobility shift analysis of cAMP-CRP binding to csrB and csrC DNA. A 32P end-labeled PCR product (0.5 nM) was used in binding reactions performed with or without cAMP (0.2 mM) and CRP, as shown. Positions of free (F) and bound (B) DNA are shown. The apparent Kd for binding of cAMP-CRP to csrC DNA was 18 ± 2 nM. Each experiment was conducted twice, and a representative image is shown.