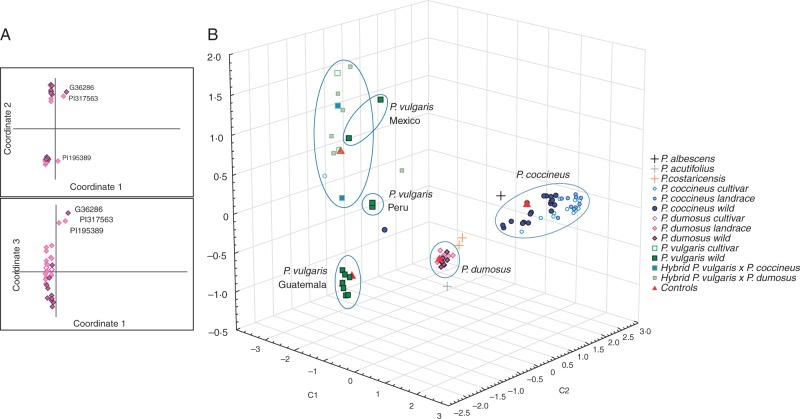
Fig. 1.
Genetic distance among species of the P. vulgaris secondary gene pool. (A) Principal coordinates analysis of 742 DArT markers for P. dumosus indicating that 72·10 % of the total genetic variation is found among the first three components. Pink diamonds, black outline, wild accessions; pink diamonds, pink outline, landraces; open diamonds, cultivars. (B) Principal coordinates analysis accounting for 72·52 % of the molecular variation found among 704 DArT markers, with 85 % of variation in P. dumosus accounted for by the first three components. P. dumosus: wild (pink diamonds, black outline), landrace (pink diamonds, pink outline), cultivar (open diamonds). P. coccineus: wild (solid dark blue circles), landrace (solid light blue circles), cultivar (open circles). P. vulgaris: wild (solid green squares), cultivar (open squares). P. costaricensis (yellow +), P. albescens (black +), P. acutifolius (grey +; outgroup). Hybrids: P. vulgaris × P. dumosus (solid pink squares), P. vulgaris × P. coccineus (solid blue squares). Controls (red triangles).