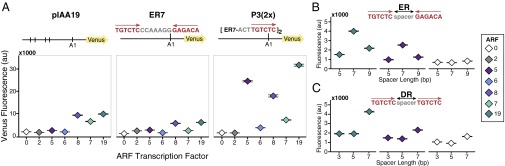
Fig. 1.
Promoter architecture can specify ARF activity. (A) ARFs are differentially active on native and synthetic promoters. Flow cytometry was used to quantify the activity of ARF transcription factors on promoter variants with variable number, spacing, and orientation of binding sites. In each subpanel, yeast strains contain the same Venus reporter construct and differ only by the ARF expressed (2, 5, 6, 8, 7, or 19). “0” indicates absence of an ARF transcription factor. Vertical lines in the first reporter schematic indicate the location of an ARF-binding site (AuxRE) in the native IAA19 promoter. Subsequent panels depict ARF activity when an ER7 or P3(2x) AuxRE variant is inserted at the A1 site in a pIAA19 reporter construct where all of the native AuxREs were mutated (Fig. S1A). All ARFs tested are expressed in yeast at least as well as ARF19 (Fig. S1B). (B) ARF19 and ARF5 exhibit optimal activity when there are seven nucleotides between a pair of AuxRE sites in an ER conformation or (C) DR conformation. When the number of tandem DR5 repeats is increased, all of the activator ARFs exhibit measurable activity (Fig. S1C). The data are shown as median values, with black horizontal bars demarcating 95% confidence intervals around the median.