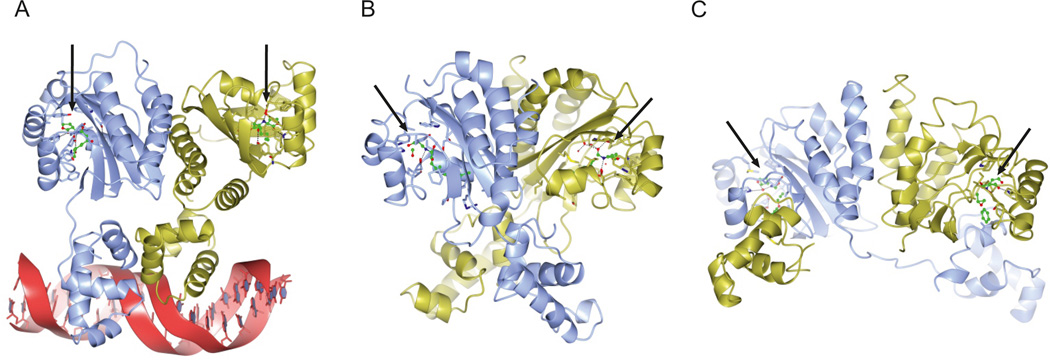
Figure 2. Structures of LuxR-type quorum sensing receptors.
This figure shows the crystal structures of four LuxR-type receptors. a | TraR from Agrobacterium tumefaciens bound to autoinducer and DNA (Protein Data Bank (PDB) entry 1L3L). b | QscR from Pseudomonas aeruginosa bound to autoinducer (PDB entry 3SZT). c | CviR from Chromobacterium violaceum bound to an inhibitor called chlorolactone (PDB entry 3QP5). The arrows denote the positions of the ligands. The structures of the ligand-binding domains of all three proteins are similar; however, whereas TraR (panel a) adopts an asymmetric dimer, QscR (panel b) and CviR (panel c) form nearly symmetric cross-subunit architectures. The locations and conformations that are adopted by the DNA-binding domains differ substantially, enabling (panels a and b) or preventing (panel c) DNA binding and transcriptional activation of target genes.