ABSTRACT
Defects in the regulation of centrosome duplication lead to tumorigenesis through abnormal cell division and resulting chromosome missegregation. Therefore, maintenance of accurate centrosome number is critical for cell fate. The deubiquitinating enzyme USP1 plays important roles in DNA repair and cell differentiation. Importantly, increased levels of USP1 are detected in certain types of human cancer, but little is known about the significance of USP1 overexpression in cancer development. Here we show that Usp1 plays a novel role in regulating centrosome duplication. The ectopic expression of wild-type Usp1, but not C90S Usp1 (catalytically inactive mutant form), induced centrosome amplification. Conversely, ablation of Usp1 in mouse embryonic fibroblasts (MEFs) showed a significant delay in centrosome duplication. Moreover, Usp1-induced centrosome amplification caused abnormal mitotic spindles, chromosome missegregation and aneuploidy. Interestingly, loss of inhibitor of DNA binding protein 1 (ID1) suppressed Usp1-induced centrosome amplification. Taken together, our results strongly suggest that Usp1 is involved in the regulation of centrosome duplication, at least in part via ID1, and Usp1 may exert its oncogenic activity, partially through inducing centrosome abnormality.
KEYWORDS: Deubiquitinating Enzymes, Usp1, Centrosome amplification, Chromosome instability, Aneuploidy
Introduction
The centrosome is the major microtubule-organizing center that controls a correct segregation of the duplicated chromosomes to daughter cells.1-3 Consequently, centrosome defects can adversely affect cell division, which contributes to the chromosome instability that is often observed during tumorigenesis.4-7 In fact, centrosome abnormalities, usually increased numbers, are common in human cancers and these changes have been observed at various stages of human cancers.6-9 Therefore, the elucidation of the regulatory mechanisms that control centrosome homeostasis is important for understanding a causal relationship between centrosome amplification and human cancers and may lead to more effective anticancer therapies.10
The USP1 gene encodes a member of the ubiquitin-specific proteases that is a deubiquitinating enzyme (DUB) with His and Cys domains.11 USP1 has been identified as a key regulator in the DNA repair processes, mainly in the Fanconi anemia pathway and Translesion DNA synthesis by regulating ubiquitination status of FANCD2 and PCNA,12,13 Mono-ubiquitinated FANCD2 and PCNA serve as a platform to recruit DNA repair proteins to DNA damage sites,14-16 and USP-induced deubiquitination of FANCD2 and PCNA is crucial for the correct function of DNA repair pathway.17,18 Indeed, Usp1 gene deletion in mice18 and USP1 inhibition by small molecules displayed chemosensitizing effects against DNA damaging agents,19-21 indicating that USP1 is required for an efficient DNA repair activity. Besides DNA repair-related function, recent study reveals that increased level of USP1 is found in human osteosarcoma cell lines.22 USP1 stabilizes inhibitor of DNA binding proteins IDs to promote the maintenance of mesenchymal stem cell in osteosarcoma, suggesting that Usp1 play an important role in cell proliferation and differentiation. The higher levels of ID proteins are evident in many tumors, implying an oncogenic role for ID proteins.23-25 Interestingly, a search of an integrated cancer microarray database (Oncomine) revealed that USP1 is overexpressed in several tumor types including cervical, gastric cancer, melanoma and sarcoma.26,27 However, little is known about the significance of USP1 overexpression in tumorigenesis.
Defects in the regulation of centrosome duplication lead to tumorigenesis through abnormal cell division and resulting inappropriate chromosome segregation. Some DUBs have been shown to have important roles in the maintenance of centrosome integrity. USP33 positively regulates centrosome duplication by stabilizing centriolar protein CP110, and USP 33 and CP110 are both upregulated in pancreatic ductal carcinomas28 in which centrosome amplification is detected.29 USP44 regulates centrosome positioning to prevent aneuploidy and suppress tumorigenesis.30 Given that increased levels of USP1 are detected in certain types of human cancer,26,27 we therefore tested if USP1 is involved in centrosome maintenance and exerts its oncogenic activity in part through centrosomal function.
In this study, we have demonstrated that ubiquitin specific protease Usp1 plays an important role in regulating centrosome duplication. The ectopic Usp1 expression in NIH3T3 cells induced centrosome amplification, whereas the ablation of Usp1 in mouse embryonic fibroblasts (MEFs) delayed centrosome duplication. Moreover, Usp1-induced centrosome amplification resulted in abnormal mitotic spindle formation, chromosome missegregation, eventually leading to aneuploidy. Furthermore, we found that ID1 is selectively required for Usp1-induced centrosome amplification. These results demonstrate for the first time that Usp1 regulates centrosome duplication and its deregulation can induce centrosome amplification, resulting in chromosome instability. Our results strongly suggest that Usp1 is directly or indirectly involved in the control of centrosome duplication, and Usp1 overexpression may contribute to oncogenic transformation, in part, by inducing centrosome abnormality and chromosome instability.
Results
Overexpression of Usp1 results in centrosome amplification
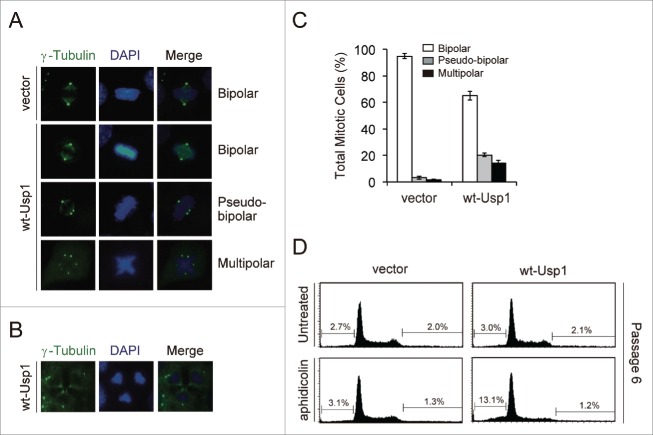
To address the effects of USP1 in centrosome duplication, NIH3T3 cells were stably transfected with empty retroviral vector or the retroviral vector encoding murine wild-type Usp1 (wt-Usp1), and the numbers of centrosomes per mononucleated cell were determined by immunofluorescence microscopy (Fig. 1). Expression of the Flag-HA-tagged exogenous Usp1 was confirmed by immunoblotting (Fig. 1A). The levels of UV-induced monoubiquitination of Fancd2 (Fancd2-Ub) and PCNA (PCNA-Ub) were reduced in wt-Usp1 expressing NIH3T3 cells compared to vector control (compare lanes 3 and 4), indicating ectopically expressed wt-Usp1 is functionally intact. Importantly, we found significantly elevated frequency of cells with extra centrosomes (centrosome amplification) in wt-Usp1 NIH3T3 cells (Fig. 1B, C). In contrast, such centrosome amplification was rarely observed in empty vector-infected cells. It has been well established that prolonged S-phase arrest leads to multiple rounds of centrosome duplication.31,32 To investigate whether the generation of extra centrosomes resulting from Usp1 overexpression is a consequence of centrosome overduplication or failure of cytokinesis, we treated NIH3T3 cells with aphidicolin (Aph) to induce prolonged S phase and compared the induction of centrosome amplification to that in untreated normally cycling cells (Fig. 1D). Usp1 overexpression triggered centrosome amplification regardless of the presence or absence of aphidicolin, so these data suggest that Usp1 overexpression causes centrosome amplification by an overduplication mechanism.
Figure 1.
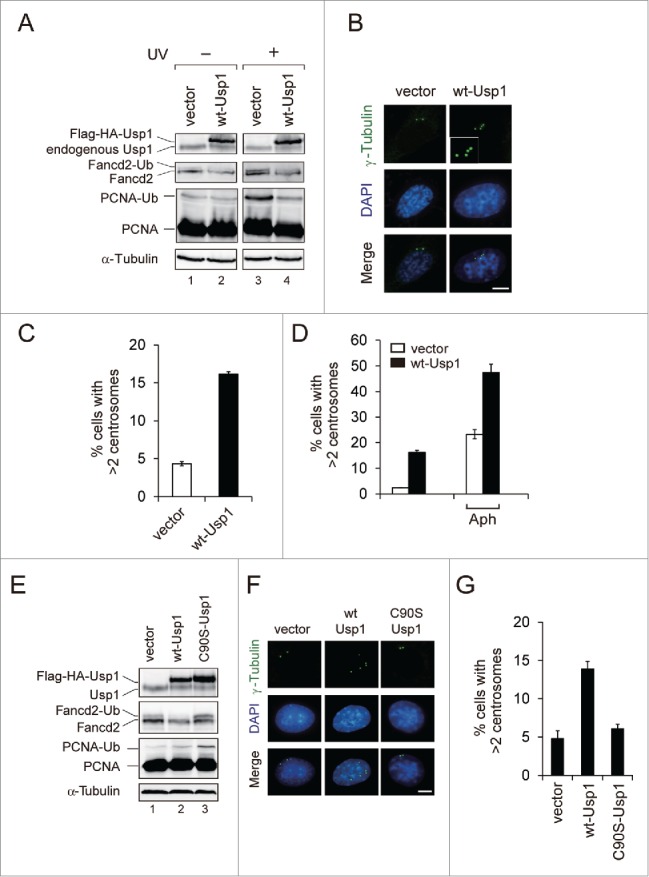
Usp1 overexpression leads to centrosome amplification. (A-D) NIH 3T3 cells were infected with retrovirus carrying wild-type Usp1 or empty vector and selected with puromycin to generate pools of resistant cells. (A) Cells were either left untreated or treated with UVC (30 J/m2, harvested at 6 hr after irradiation). Cell lysates were immunoblotted with indicated antibodies. (B) Cells were stained with anti γ-tubulin antibody (green) and DAPI (blue). Bar, 10 μm. Representative fields are shown. Inset shows high-magnification views of centrosomes in wt-Usp1 NIH3T3 cells. (C) Quantitation of results shown in (B). Histograms showing percentage of cells with extra centrosomes (more than 2 centrosomes per mononucleated cells; >2). Cumulative data from 3 independent experiments with at least 300 cells counted per experiments. Values represent the mean ± SEM. (D) Histograms showing percentages of cells with extra centrosomes (>2) incubated without (−) or with aphidicolin (2 μg/ml for 48 hr). Cumulative data from 3 independent experiments with at least 200 cells counted per experiments. Values represent the mean ± SEM. (E-G) NIH 3T3 cells were infected with retrovirus carrying C90S Usp1 and selected with puromycin to generate pools of resistant cells. (E) Cell lysates were immunoblotted with indicated antibodies. (F) Cells were stained with anti γ-tubulin antibody (green) and DAPI (blue). Bar, 10 μm. Representative fields are shown. (G) Histograms showing percentage of cells with extra centrosomes (>2). Cumulative data from 3 independent experiments with at least 200 cells counted per experiments. Values represent the mean ± SEM.
Deubiquitinating activity of Usp1 is required for centrosome amplification
To investigate whether the induction of centrosome amplification by Usp1 depends on its deubiquitinating activity (Fig. 1E-G), we generated NIH3T3 cells expressing catalytically inactive Usp1 mutant (C90S-Usp1). Immunoblotting for Fancd2 and PCNA revealed elevated monoubiquitination in C90S-Usp1 NIH3T3 cells (Fig. 1E; compare lanes 1 and 3), indicating that C90S-Usp1 might compete with endogenous Usp1 leading to inhibition of endogenous Usp1 activity. Importantly, C90S-Usp1 failed to induce centrosome amplification in NIH3T3 cells (Fig. 1F, G). This result indicates that deubiquitinating activity of Usp1 is required for its ability to induce centrosome amplification.
Usp1 ablation delays centrosome duplication
The centrosome duplication initiates at the G1/S transition and is completed during S phase in mammalian cells.1 USP1 protein levels are downregulated during the G1 phase, begin to accumulate as cells entered S phase and remain elevated until the late stages of mitosis.33,34 To examine whether loss of Usp1 function can cause a defect in centrosome duplication during S phase, we measured the centrosome number from immortalized Usp1+/+ and Usp1−/− MEFs (Fig. 2A-C). As reported previously,18 increased levels of Fancd2-Ub and PCNA-Ub were detected in Usp1−/− MEFs (Fig. 2A). Usp1+/+ and Usp1−/− MEFs showed similar percentage of S phase cells and growth rates, indicating that there was no apparent difference in the cell cycle progression between Usp1+/+ and Usp1−/− MEFs (Fig. 2B and Fig. S2A). Under the condition examined, about 82% of Usp1+/+ MEFs contained 2 centrosome per cell, with ∼14 % of the cells containing a single centrosome. In contrast, only 44 % of Usp1−/− MEFs, however, contained duplicated centrosomes, with ∼53 % of the cells containing a single centrosome (Fig. 2C), indicating that centrosome duplication is defective in Usp1−/− MEFs without affecting cell-cycle distribution. To further address the role of USP1 in centrosome duplication, we investigated whether USP1 depletion could affect centrosome overduplication in U2OS cells treated with aphidicolin (Fig. 2D-F). Acute depletion of USP1 by siRNA did not change cell cycle distribution (Fig. 2D). Importantly, depletion of USP1 suppressed aphidicolin-induced centrosome amplification (Fig. 2E, F), suggesting that centrosome phenotype provoked by USP1 silencing was not due to altered cell-cycle progression and USP1 might be involved in the regulation of centrosome duplication.
Figure 2.
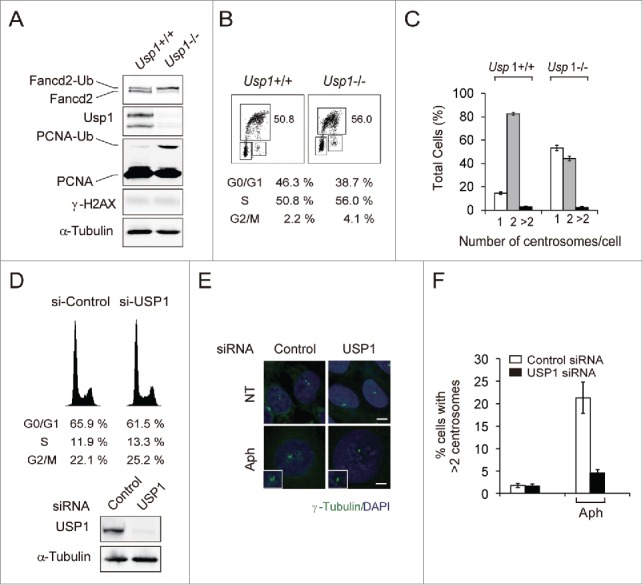
Usp1−/− MEFs have delayed centrosome duplication. (A) Immortalized Usp1+/+ and Usp1−/− MEFs were processed for immunoblotting with the indicated antibodies. (B) Cell cycle distribution of Usp1+/+ and Usp1−/− MEFs as determined by BrdU incorporation. The percentages of cell populations of different cell cycle phases are shown. Representative data are shown. (C) Quantification of centrosome number in Usp1+/+ and Usp1−/− MEFs using γ-tubulin labeling. Cumulative data from 3 independent experiments with at least 200 interphase cells counted per experiments. Values represent the mean ± SEM. (D-F) U2OS cells transfected with indicated siRNA for 24 hr were incubated for a further 48hr without or with aphidicolin (2 μg/ml). (D) U2OS cells transfected with indicated siRNA for 72hr were processed for FACS analysis (upper panel) and immnoblotting (lower panel). (E) Cells were stained with anti γ-tubulin antibody (green) and DAPI (blue). Bar, 10 μm. Representative fields are shown. (F) Histograms showing percentage of cells with extra centrosomes (>2). Cumulative data from 3 independent experiments with at least 200 cells counted per experiments. Values represent the mean ± SEM.
Usp1-induced centrosome amplification causes abnormal spindle formation and chromosome missegregation
Given that deletion of Usp1 delays normal centrosome duplication and that its overexpression causes centrosome amplification, an important question was whether Usp1-induced centrosome amplification has any pathological consequences. We therefore examined whether Usp1 overexpression affects spindle formation and chromosome segregation during mitosis (Fig. 3A-C). Notably, 34 % of dividing wt-Usp1 NIH3T3 cells showed abnormal spindles with extra centrosomes (Fig. 3C). In these cells showing abnormal spindle formation, 59% of cells displayed pseudo-bipolar spindles (termed “centrosome clustering”,35 and 41% of cells displayed multipolar spindles. Occasionally, some cells showing multipolar spindles were seen to undergo chaotic cytokinesis with triple midbody (Fig. 3B), which perhaps can lead to the generation of aneuploid cells.
Figure 3.
Usp1-induced centrosome amplification results in abnormal spindle formation, chromosome missegregation and aneuploidy. (A) Representative mitotic cells with bipolar or multipolar spindle poles are shown. (B) A representative image for wt-Usp1 NIH3T3 cells with 3-directional cell division. (C) Histogram showing percentages of mitotic cells with bipolar or multipolar spindle poles in vector- and wt-Usp1 NIH3T3 cells. Cumulative data from 3 independent experiments with at least 100 mitotic cells counted per experiments. Values represent the mean ± SEM. (D) The vector- and wt-Usp1 NIH3T3 were incubated without or with aphidicolin (5 μg/ml) for 24 hr and cultured for the indicated number of passages (every 3 days) and then analyzed for DNA content by flow cytometry. Representative histogram plots and the percentage of aneuploid cells are shown.
Usp1-induced centrosome amplification results in chromosome instability and aneuploidy
Since aneuploidy is a cellular consequence that is often associated with centrosome amplification and mitotic spindle defects,4,36 we next examined whether Usp1-induced centrosome amplification generates aneuploid cells during long-term serial passage. However, we did not observe significant differences between normally cycling vector- and wt-Usp1 NIH 3T3 cells (Fig. 3D; upper panels). To increase the frequency of extra centrosomes in wt-Usp1 NIH 3T3 cells, cells were treated with aphidicolin and cultured for the indicated number of passages. To measure the percentage of aneuploid cells by FACS analysis, sub-G1 population undergoing apoptosis or necrosis were initially excluded. In passage 1 (P1), wt-Usp1 NIH3T3 cells showed a similar cell-cycle distribution to the vector NIH 3T3 cells (data not shown). However, following 5 passages (P6), significant increase in aneuploidy in sub-G1 phase were observed in wt-Usp1 NIH3T3 cells (Fig. 3D; lower panels). In contrast, Aph-treated vector NIH 3T3 cells did not show any increases in sub-G1 population at P6, suggesting that abnormal spindle formation in wt-Usp1 NIH3T3 cells may generate aneuploid daughter cells.
Overexpression of wt Usp1, but not C90S Usp1, increases the levels of ID1 and ID2
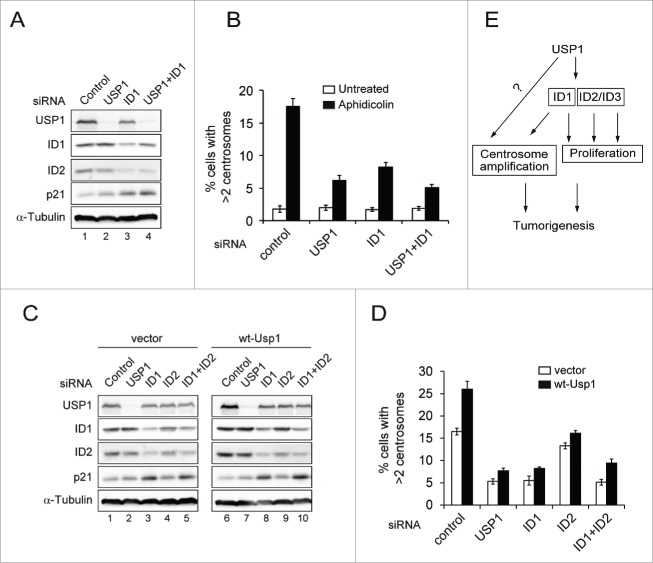
It has been reported that USP1 stabilizes 3 members of the family of ID proteins through deubiquitination.22 Interestingly, ID1, but not other ID proteins (ID2, ID3 and ID4), is localized at centrosome and forced expression of ID1 results in accumulation of supernumerary centrosomes,37-39 suggesting that stabilization of ID1 through USP1 overexpression can induce centrosome amplification. To explore this possibility, we generated NIH3T3 cells stably transduced with Id1 and compared the induction of centrosome amplification with wt-Usp1 NIH3T3 cells (Fig. 4A and B). As expected, we observed a significant stabilization of Id1 and Id2 in wt-Usp1 NIH3T3 cells (Fig. 4A and Fig. 4C-E). Interestingly, despite higher expression of Id1 in Id1-NIH3T3 cells, total frequency of centrosome amplification was lower than that of wt-Usp1-NIH3T3 cells (Fig. 4B), suggesting that additional factors might be involved in Usp1-induced centrosome amplification.
Figure 4.
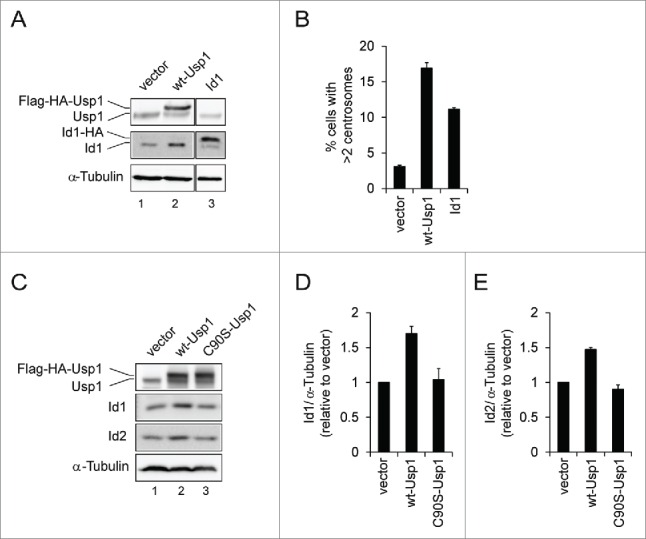
Overexpression of wt-Usp1, but not C90S-Usp1, stabilizes the levels of ID1 and ID2 (A-B) NIH3T3 cells were infected with retrovirus carrying murine Id1 and selected with puromycin to generate pools of resistant cells. (A) Cell lysates were immunoblotted with indicated antibodies. (B) Histograms showing percentage of cells with extra centrosomes (>2). Cumulative data from 3 independent experiments with at least 150 cells counted per experiments. Values represent the mean ± SEM. (C-E) Quantification of the ratio of ID1 to α-tubulin and ID2 to α-tubulin in vector-, wt-Usp1, and C90S-Usp1 NIH3T3 cells. Representative example of immunoblots analysis of NIH3T3 cells (C). The ratio of ID1/α-tubulin (D) and ID2/α-tubulin (E) was determined by densitometry analysis of the immunoblot bands. The values represent the mean ± SEM of 3 independent experiments.
Usp1 induces centrosome amplification, at least in part, via inducing ID1 expression
To explore the mechanism by which Usp1 overexpression impacts on centrosome duplication, USP1/ID1 co-depleted cells were compared to cells individually depleted of either USP1 or ID1 (Fig. 5A and B). It has been shown that shRNA-mediated USP1 depletion reduced the levels of ID1 and ID2, resulting in p21-induced cell-cycle arrest.22 However, acute depletion of USP1 did not affect the levels of ID1 and ID2 in U2OS cells (Fig. 5A), this discrepancy could be explained by different depletion methods. Depletion of either USP1 or ID1 inhibited Aph-induced centrosome amplification (Fig. 5B). Importantly, co-depletion of USP1/ID1 showed a further decrease in Aph-induced centrosome amplification compared with depletion of either USP1 or ID1, although the differences were very small (Fig. 5B). To further assess the contribution of the ID1 to Usp1-induced centrosome amplification, we generated U2OS cells stably transduced with wt-Usp1 and investigated whether depletion of ID1 could affect Usp1-induced centrosome amplification (Fig. 5C-D and Fig. S3B). Similar to wt-Usp1 NIH3T3 cells, U2OS cells expressing wt-Usp1 displayed the higher frequency of Aph-induced centrosome amplification compared to empty vector-transduced cells (Fig. 5C). Importantly, depletion of ID1 markedly reduced centrosome amplification in wt-Usp1-U2OS cells, whereas depletion of ID2 slightly affected, indicating that the ID1 is selectively required for USP1-induced centrosome amplification. Moreover, after combined depletion of ID1 & ID2, there was no further reduction of centrosome amplification. However considering the depletion efficiency of ID2 (Fig. 5C), we cannot exclude the possibility that partial effect by ID2 depletion could be attributable to inefficient ID2 depletion.
Figure 5.
Usp1 induces centrosome amplification at least in part through ID1 (A-B) U2OS cells transfected with indicated siRNA for 24hr were incubated for a further 48hr without or with aphidicolin (2 μg/ml). (A) U2OS cells transfected with indicated siRNA for 72hr were analyzed with indicated antibodies (B) Cells were stained with anti γ-tubulin antibody. Histograms showing percentage of cells with extra centrosomes (>2). Cumulative data from 3 independent experiments with at least 200 cells counted per experiments. Values represent the mean ± SEM. (C-D) U2OS cells stably transduced with empty vector or wt-Usp1 were transfected with indicated siRNA for 24hr and incubated for a further 48hr with aphidicolin (2 μg/ml). (C) The vector-U2OS and wt-Usp1 U2OS cells transfected with indicated siRNA for 72 hr were analyzed with indicated antibodies (D) Histograms showing percentage of cells with extra centrosomes (>2). Cumulative data from 3 independent experiments with at least 200 cells counted per experiments. Values represent the mean ± SEM. (E) A proposed model for how USP1 exerts its oncogenic properties in tumorigenesis.
Taken together, our findings indicate that Usp1 overexpression induces centrosome amplification and chromosomal instability, and suggest that the role of Usp1 in centrosome duplication, at least in part through ID1, may be partially responsible for its oncogenic property (Fig. 5E).
Discussion
We have demonstrated that ubiquitin specific protease Usp1 plays an important role in regulating centrosome duplication. The overexpression of Usp1 in NIH 3T3 cells induces centrosome amplification in both normally cycling and S phase-arrest conditions (Fig. 1C, D). Importantly, the ability of Usp1 to induce centrosome amplification is abolished by point mutations that disrupt Usp1 activity (Fig. 1G). Conversely, Usp1-deficient MEFs showed a significant delay in centrosome duplication without affecting cell-cycle distribution (Fig. 2C). Moreover, silencing of USP1 inhibited aphidicolin-induced centrosome amplification (Fig. 2F and Fig. 5B). Furthermore, Usp1-induced centrosome amplification caused abnormal mitotic spindles, chromosome missegregation, eventually leading to aneuploidy (Fig. 3). Lastly, loss of ID1 significantly inhibited Usp1-induced centrosome amplification after treatment of aphidicolin (Fig. 5D).
How Usp1 regulates centrosome duplication and how its deregulation leads to centrosome amplification remain to be defined, and several models are possible. i) Recent studies have shown that several DNA repair proteins localize to the centrosome, and defects in these proteins cause several functional aberrations in centrosomes such as centrosome amplification.40-43 Also, it has been shown that DNA damage can induce centrosome amplification.41-43 Overexpression of Usp1 in NIH3T3 cells inhibited UV-induced Fancd2-Ub and PCNA-Ub (Fig. 1A). Since mono-ubiquitinated forms of FANCD2 and PCNA recruit DNA repair proteins to DNA damage sites,12,15 one might expect Usp1 overexpression could accumulate DNA damage in wt-Usp1 NIH3T3 cells, resulting in centrosome amplification. However, we did not observe any differences in γ-H2AX and CHK1 phosphorylation between vector-and wt-Usp1 NIH3T3 cells, suggesting that there were no indications of spontaneous DNA damage (Fig. S1C, D). ii) Alternatively, USP1 has been reported to deubiquitinate and stabilize ID proteins.22 Overexpression of USP1 led to an increase in ID1, ID2 and ID3 abundance and cell proliferation. Interestingly, ID1, but not ID2 and ID3, is localized at centrosome and ID1 overexpression results in accumulation of supernumerary centrosomes.37-39 Our findings suggest the possibility for the involvement of ID1 in USP1-induced centrosome amplification. Indeed, an increase in Id1 abundance was detected in wt-Usp1 NIH3T3 cells (Fig. 4). Notably, ID1 depletion in USP1-expressing U2OS cells showed great reduction in the frequency of centrosome amplification, whereas ID2 depletion showed only modest reduction (Fig. 5C, D). Our results suggest that stabilization of ID1, but not ID2, by USP1, may selectively contribute to USP1-induced centrosome amplification. iii) Lastly, although centrosomal localization of Usp1 has not been detected yet, we cannot completely exclude the possibility that Usp1 may directly interact and modify the centrosomal proteins.
Usp1−/− MEFs showed a significant delay in centrosome duplication. Moreover, depletion of Usp1 in U2OS cells suppressed centrosome amplification induced by aphidicolin. One might expect that genome instability in Usp1−/− MEFs may cause cell-cycle defects leading to delayed centrosome duplication. However, there were no observable cell cycle defects and genome instability in the absence of DNA damaging agents in Usp1−/− MEFs (Fig. 2B and Fig. S2). We observed significantly reduced level of ID1 in Usp1−/− MEFs (Fig. S3A), suggesting that Usp1 might be involved in the regulation of centrosome duplication, at least in part via ID1. However, the reduction of ID1 level was not observed in USP1-depleted U2OS cells where Aph-induced centrosome amplification was suppressed without affecting ID1 level (Fig. 5A-D). Therefore, further investigation will be required to understand how Usp1 is involved in regulation of centrosome duplication and to identify possible candidates.
The centrosome amplification leads to tumorigenesis through abnormal cell division and resulting chromosome instability.4,7,36,44,45 Thus, it would be interesting to investigate whether elevated levels of USP1 correlates with increased centrosome abnormalities in human cancers. In summary, we have demonstrated for the first time that Usp1 plays a novel role in regulating centrosome duplication. Our findings strongly suggest that Usp1-induced centrosome abnormalities may contribute to chromosome instability and oncogenic transformation (Fig. 5E). These results also suggest that inhibition of USP1 might provide a new therapeutic strategy to inhibit centrosome amplification and chromosome instability in cancer cells.
Materials and Methods
Cell culture and chemical
Usp1+/+ and Usp1−/− MEFs were described previously.18 Usp1+/+, Usp1−/− MEFs, NIH3T3 and U2OS cells were maintained in DMEM supplemented with 10% fetal calf serum. Aphidicolin was from Sigma.
Antibodies
Rabbit anti-Usp1 and anti-Fancd2 antibodies were described previously.18 Other antibodies used in this study were as follows; anti-PCNA, anti-ID1 and anti-ID2 antibodies (Santa Cruz Biotechnology), anti-α-Tubulin and anti-γ-tubulin antibodies (Sigma-Aldrich), anti-BrdU antibody (BD PharMingen), anti-γ-H2AX antibody (Millipore), anti-CHK1 S345 and S317 antibodies (Cell Signaling Technology), anti-HA antibody (Roche Life Science). Secondary antibodies were peroxidase-conjugated anti-rabbit or anti-mouse (Cell Signaling Technology).
SiRNAs transfection
The target sequences of USP1, ID1 and ID2 are aaccagagacaaactagatca, ctccagcacgtcatcgactac and gagttaatgtcaaatgacagc, respectively. Control siRNA is designed by Qiagen. SiRNA transfection was performed with RNAimax (Invitrogen).
Retrovirus infection
The wild-type murine Usp1 cDNA was sub-cloned into the retroviral vector pFLAG-HA-MSCV-puro. The Usp1C90S and Usp1GG/AA mutants were generated by Site-directed Mutagenesis kit (Stratagene). Each Usp1 construct was transfected into 293T cells with packaging plasmid by using Lipofectamine 3000 (Invitrogen). After 48 and 60 hr from DNA transfection, immortalized MEFs, NIH3T3 cells and U2OS cells were infected with retroviral supernatant in the presence of 5 μg/ml polybrene and were allowed to grow for 2 more days. The infected cells were selected with 3 μg/ml puromycin and viable cells were expanded.
Immunofluorescence
To detect centrosomes, cells were fixed with ice-cold methanol for 10 min at −20°C and permeabilized with 0.5% Triton X-100 for 5 min. To detect γ-H2AX foci, cells were pre-extracted with 0.2% Triton X-100 for 1 min, followed by fixation with 3.7% paraformaldehyde for 10 min and permeabilized. Cells were blocked with 3% BSA/PBS and were incubated with either anti γ-tubulin antibody (1:5000) or anti γ-H2AX antibody (1:2000) for 1h. Alexa 488-conjugated secondary antibody (Invitrogen) was used at a dilution of 1:500. Coverslips were mounted in a DAPI-containing mounting medium (Molecular Probes) and analyzed.
Detection of aneuploid cells by FACS analysis
Cells were trypsinized, fixed with cold 70% ethanol, treated with propidium iodide and ribonuclease A, and subjected to cell cycle analysis using FACS calibur (Becton Dickinson). Percentage of aneuploid cells was calculated with ModFit LT cell-cycle analysis software.
BrdU incorporation
Usp1+/+, Usp1−/− MEFs, NIH3T3 cells were incubated for 1 hour with 10 μM BrdU (BD PharMingen). After fixation with 70% ethanol, the cells were stained with FITC conjugated anti-BrdU antibody, and counterstained with propidium iodide, according to the manufacturer's instruction (Becton Dickinson).
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We especially would like to thank A. D’Andrea for valuable tools and reagents for this study. We also thank to Mira Kim who was involved in the initial stages of the project, and I Park, KK Kim, N Kim, and H Kook for helpful discussions.
Author contributions
J.K.J and S.W.J conducted overall experiments; J.M.K. designed the experiments and wrote the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (Medical Research Council for Gene Regulation) funded by the Ministry of Science, ICT and Future Planning (2011-0030132). This work was supported in part by Basic science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (2014R1A1A2054983).
References
- [1].Doxsey S, Zimmerman W, Mikule K. Centrosome control of the cell cycle. Trends Cell Biol 2005; 15:303-11; PMID:15953548; http://dx.doi.org/ 10.1016/j.tcb.2005.04.008 [DOI] [PubMed] [Google Scholar]
- [2].Doxsey S, McCollum D, Theurkauf W. Centrosomes in cellular regulation. Annu Rev Cell Dev Biol 2005; 21:411-34; PMID:16212501; http://dx.doi.org/ 10.1146/annurev.cellbio.21.122303.120418 [DOI] [PubMed] [Google Scholar]
- [3].Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol 2002; 14:25-34; PMID:11792541; http://dx.doi.org/ 10.1016/S0955-0674(01)00290-3 [DOI] [PubMed] [Google Scholar]
- [4].Vitre BD, Cleveland DW. Centrosomes, chromosome instability (CIN) and aneuploidy. Curr Opin Cell Biol 2012; 24:809-15; PMID:23127609; http://dx.doi.org/ 10.1016/j.ceb.2012.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature 2009; 460:278-82; PMID:19506557; http://dx.doi.org/ 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer 2002; 2:815-25; PMID:12415252; http://dx.doi.org/ 10.1038/nrc924 [DOI] [PubMed] [Google Scholar]
- [7].D'Assoro AB, Lingle WL, Salisbury JL. Centrosome amplification and the development of cancer. Oncogene 2002; 21:6146-53; PMID:12214243; http://dx.doi.org/ 10.1038/sj.onc.1205772 [DOI] [PubMed] [Google Scholar]
- [8].Chan JY. A clinical overview of centrosome amplification in human cancers. Int J Biol Sci 2011; 7:1122-44; PMID:22043171; http://dx.doi.org/ 10.7150/ijbs.7.1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell 2009; 139:663-78; PMID:19914163; http://dx.doi.org/ 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- [10].Korzeniewski N, Hohenfellner M, Duensing S. The centrosome as potential target for cancer therapy and prevention. Expert Opin Ther Targets 2013; 17:43-52; PMID:23062185; http://dx.doi.org/ 10.1517/14728222.2013.731396 [DOI] [PubMed] [Google Scholar]
- [11].Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, Bernards R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell 2005; 17:331-9; PMID:15694335; http://dx.doi.org/ 10.1016/j.molcel.2005.01.008 [DOI] [PubMed] [Google Scholar]
- [12].Huang TT, D'Andrea AD. Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol 2006; 7:323-34; PMID:16633336; http://dx.doi.org/ 10.1038/nrm1908 [DOI] [PubMed] [Google Scholar]
- [13].Kim H, D'Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev 2012; 26:1393-408; PMID:22751496; http://dx.doi.org/ 10.1101/gad.195248.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D'Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol 2006; 8:339-47; PMID:16531995 [DOI] [PubMed] [Google Scholar]
- [15].Kee Y, D'Andrea AD. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev 2010; 24:1680-94; PMID:20713514; http://dx.doi.org/ 10.1101/gad.1955310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer 2003; 3:23-34; PMID:12509764; http://dx.doi.org/ 10.1038/nrc970 [DOI] [PubMed] [Google Scholar]
- [17].Oestergaard VH, Langevin F, Kuiken HJ, Pace P, Niedzwiedz W, Simpson LJ, Ohzeki M, Takata M, Sale JE, Patel KJ. Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol Cell 2007; 28:798-809; PMID:18082605; http://dx.doi.org/ 10.1016/j.molcel.2007.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kim JM, Parmar K, Huang M, Weinstock DM, Ruit CA, Kutok JL, D'Andrea AD. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell 2009; 16:314-20; PMID:19217432; http://dx.doi.org/ 10.1016/j.devcel.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mistry H, Hsieh G, Buhrlage SJ, Huang M, Park E, Cuny GD, Galinsky I, Stone RM, Gray NS, D'Andrea AD, et al.. Small-molecule inhibitors of USP1 target ID1 degradation in leukemic cells. Mol Cancer Ther 2013; 12:2651-62; PMID:24130053; http://dx.doi.org/ 10.1158/1535-7163.MCT-13-0103-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liang Q, Dexheimer TS, Zhang P, Rosenthal AS, Villamil MA, You C, Zhang Q, Chen J, Ott CA, Sun H, et al.. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat Chem Biol 2014; 10:298-304; PMID:24531842; http://dx.doi.org/ 10.1038/nchembio.1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chen J, Dexheimer TS, Ai Y, Liang Q, Villamil MA, Inglese J, Maloney DJ, Jadhav A, Simeonov A, Zhuang Z. Selective and cell-active inhibitors of the USP1/ UAF1 deubiquitinase complex reverse cisplatin resistance in non-small cell lung cancer cells. Chem Biol 2011; 18:1390-400; PMID:22118673; http://dx.doi.org/ 10.1016/j.chembiol.2011.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Williams SA, Maecker HL, French DM, Liu J, Gregg A, Silverstein LB, Cao TC, Carano RA, Dixit VM. USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell 2011; 146:918-30; PMID:21925315; http://dx.doi.org/ 10.1016/j.cell.2011.07.040 [DOI] [PubMed] [Google Scholar]
- [23].Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol 2003; 13:410-8; PMID:12888293; http://dx.doi.org/ 10.1016/S0962-8924(03)00147-8 [DOI] [PubMed] [Google Scholar]
- [24].Sikder HA, Devlin MK, Dunlap S, Ryu B, Alani RM. Id proteins in cell growth and tumorigenesis. Cancer Cell 2003; 3:525-30; PMID:12842081; http://dx.doi.org/ 10.1016/S1535-6108(03)00141-7 [DOI] [PubMed] [Google Scholar]
- [25].Norton JD. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci 2000; 113 ( Pt 22):3897-905; PMID:11058077 [DOI] [PubMed] [Google Scholar]
- [26].Luise C, Capra M, Donzelli M, Mazzarol G, Jodice MG, Nuciforo P, Viale G, Di Fiore PP, Confalonieri S. An atlas of altered expression of deubiquitinating enzymes in human cancer. PloS One 2011; 6:e15891; PMID:21283576; http://dx.doi.org/ 10.1371/journal.pone.0015891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Garcia-Santisteban I, Peters GJ, Giovannetti E, Rodriguez JA. USP1 deubiquitinase: cellular functions, regulatory mechanisms and emerging potential as target in cancer therapy. Mol Cancer 2013; 12:91; PMID:23937906; http://dx.doi.org/ 10.1186/1476-4598-12-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Li J, D'Angiolella V, Seeley ES, Kim S, Kobayashi T, Fu W, Campos EI, Pagano M, Dynlacht BD. USP33 regulates centrosome biogenesis via deubiquitination of the centriolar protein CP110. Nature 2013; 495:255-9; PMID:23486064; http://dx.doi.org/ 10.1038/nature11941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sato N, Mizumoto K, Nakamura M, Nakamura K, Kusumoto M, Niiyama H, Ogawa T, Tanaka M. Centrosome abnormalities in pancreatic ductal carcinoma. Clin Cancer Res 1999; 5:963-70; PMID:10353727 [PubMed] [Google Scholar]
- [30].Zhang Y, Foreman O, Wigle DA, Kosari F, Vasmatzis G, Salisbury JL, van Deursen J, Galardy PJ. USP44 regulates centrosome positioning to prevent aneuploidy and suppress tumorigenesis. J Clin Invest 2012; 122:4362-74; PMID:23187126; http://dx.doi.org/ 10.1172/JCI63084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Balczon R, Bao L, Zimmer WE, Brown K, Zinkowski RP, Brinkley BR. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J Cell Biol 1995; 130:105-15; PMID:7790366; http://dx.doi.org/ 10.1083/jcb.130.1.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat Cell Biol 1999; 1:88-93; PMID:10559879; http://dx.doi.org/ 10.1038/10054 [DOI] [PubMed] [Google Scholar]
- [33].Cotto-Rios XM, Jones MJ, Huang TT. Insights into phosphorylation-dependent mechanisms regulating USP1 protein stability during the cell cycle. Cell Cycle 2011; 10:4009-16; PMID:22101265; http://dx.doi.org/ 10.4161/cc.10.23.18501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cotto-Rios XM, Jones MJ, Busino L, Pagano M, Huang TT. APC/CCdh1-dependent proteolysis of USP1 regulates the response to UV-mediated DNA damage. J Cell Biol 2011; 194:177-86; PMID:21768287; http://dx.doi.org/ 10.1083/jcb.201101062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev 2008; 22:2189-203; PMID:18662975; http://dx.doi.org/ 10.1101/gad.1700908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Dey P. Aneuploidy and malignancy: an unsolved equation. J Clin Pathol 2004; 57:1245-9; PMID:15563660; http://dx.doi.org/ 10.1136/jcp.2004.018952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hasskarl J, Duensing S, Manuel E, Munger K. The helix-loop-helix protein ID1 localizes to centrosomes and rapidly induces abnormal centrosome numbers. Oncogene 2004; 23:1930-8; PMID:14755252; http://dx.doi.org/ 10.1038/sj.onc.1207310 [DOI] [PubMed] [Google Scholar]
- [38].Man C, Rosa J, Yip YL, Cheung AL, Kwong YL, Doxsey SJ, Tsao SW. Id1 overexpression induces tetraploidization and multiple abnormal mitotic phenotypes by modulating aurora A. Mol Biol Cell 2008; 19:2389-401; PMID:18353975; http://dx.doi.org/ 10.1091/mbc.E07-09-0875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang X, Di K, Zhang X, Han HY, Wong YC, Leung SC, Ling MT. Id-1 promotes chromosomal instability through modification of APC/C activity during mitosis in response to microtubule disruption. Oncogene 2008; 27:4456-66; PMID:18372912; http://dx.doi.org/ 10.1038/onc.2008.87 [DOI] [PubMed] [Google Scholar]
- [40].Nalepa G, Enzor R, Sun Z, Marchal C, Park SJ, Yang Y, Tedeschi L, Kelich S, Hanenberg H, Clapp DW. Fanconi anemia signaling network regulates the spindle assembly checkpoint. J Clin Invest 2013; 123:3839-47; PMID:23934222; http://dx.doi.org/ 10.1172/JCI67364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Dodson H, Wheatley SP, Morrison CG. Involvement of centrosome amplification in radiation-induced mitotic catastrophe. Cell Cycle 2007; 6:364-70; PMID:17297293; http://dx.doi.org/ 10.4161/cc.6.3.3834 [DOI] [PubMed] [Google Scholar]
- [42].Hut HM, Lemstra W, Blaauw EH, Van Cappellen GW, Kampinga HH, Sibon OC. Centrosomes split in the presence of impaired DNA integrity during mitosis. Mol Biol Cell 2003; 14:1993-2004; PMID:12802070; http://dx.doi.org/ 10.1091/mbc.E02-08-0510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Inanc B, Morrison CG. Getting permission: how DNA damage causes centrosome amplification. Cell Cycle 2011; 10:1890-1; PMID:21673504; http://dx.doi.org/ 10.4161/cc.10.12.15648 [DOI] [PubMed] [Google Scholar]
- [44].Pihan GA. Centrosome dysfunction contributes to chromosome instability, chromoanagenesis, and genome reprograming in cancer. Front Oncol 2013; 3:277; PMID:24282781; http://dx.doi.org/ 10.3389/fonc.2013.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Vitale I, Galluzzi L, Castedo M, Kroemer G. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol 2011; 12:385-92; PMID:21527953; http://dx.doi.org/ 10.1038/nrm3115 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.