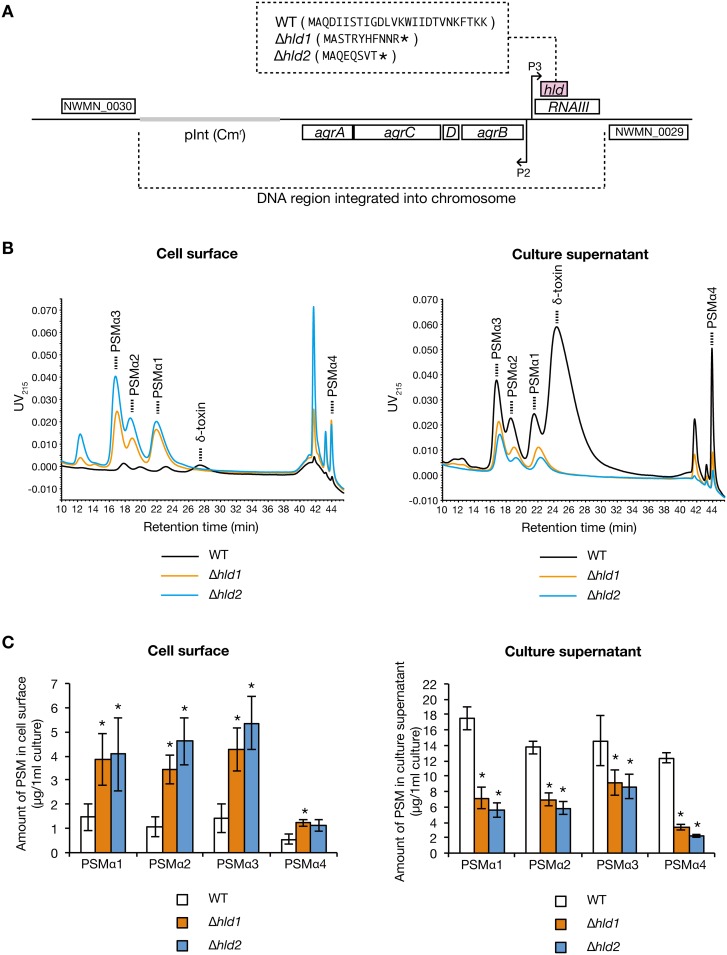
Fig 4. Effect of δ-toxin knockout on PSMαs distribution.
A. Schematic representation of genomic region of hld-wild-type strain (WT) and two different δ-toxin knockout strains (Δhld1, Δhld2) of S. aureus Newman strain. An integration vector carrying the agr locus having wild-type hld gene or mutated hld genes was integrated into the intergenic region between NWMN_0029 and NWMN_0030 genes in the chromosome of the agr null mutant of Newman strain [18]. Hld amino acid sequences are presented in the parentheses. Asterisks in the parentheses indicate stop codons resulted from artificial point mutation [18]. The regulatory function of the agr locus is the same between WT, Δhld1, and Δhld2 [18]. B. S. aureus hld-wild-type strain (WT) and two different δ-toxin knockout strains (Δhld1, Δhld2) were cultured for 19 h. PSMs on the cell surface (from 1.33 ml bacterial culture) and in the culture supernatant (from 0.267 ml bacterial culture) were analyzed by HPLC. Dotted line represents each PSM. C. Amount of PSMα1, PSMα2, PSMα3, and PSMα4 on the cell surface and in the culture supernatant of the hld-wild-type stain and the δ-toxin knockout strains were measured. Vertical axis represents the amount of each PSM per 1 ml bacterial culture. Data are means±standard errors from four independent experiments. Asterisks indicate Student’s t-test p value less than 0.05 between WT and Δhld1 or between WT and Δhld2.