Figure 1.
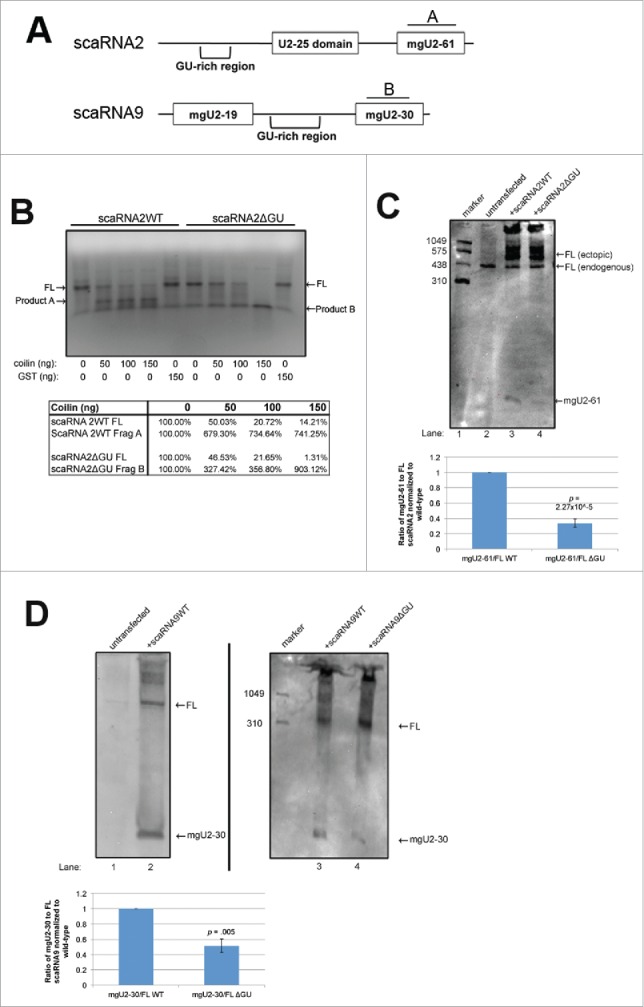
Deletion of GU-rich regions in scaRNA2 and 9 results in a decrease in processing. (A) Schematic of scaRNA2 and scaRNA9 used in subsequent experiments. A and B indicate the binding location for probes used in Northern detection. (B) RNA degradation assay using scaRNA2 wildtype (WT) and scaRNA2 with its GU-rich region deleted (ΔGU). RNAs were treated with either no protein; 10ng, 100ng, 150ng bacterially purified coilin; or 150ng GST as a control. The location of full-length (FL) scaRNA2 or scaRNA2ΔGU is indicated, as are their degradation products (A for scaRNA2 and B for scaRNA2ΔGU). The quantification of this data is shown below gel. (C,D) ScaRNA2WT, scaRNA2ΔGU, scaRNA9, or scaRNA9ΔGU pcDNA 3.1+ constructs were expressed in HeLa cells for 24 hours. Isolated RNA was then subjected to Northern blotting and probing with probe A (for scaRNA2 RNAs) or probe B (for scaRNA9 RNAs). The mgU2-61 signal for scaRNA2 is significantly reduced in ΔGU expressing cells compared to WT (n = 3 experimental repeats, p = 0.005) (histogram). The mgU2-30 signal for scaRNA9 is significantly reduced in ΔGU expressing cells compared to WT (n = 3 experimental sets, p = 2.27×10−5) (histogram). For panel D, scaRNA9 can be observed in untransfected cells, but the mgU2-30 fragment is difficult to detect (lane 1).
