ABSTRACT
American Foulbrood Disease, caused by the bacterium Paenibacillus larvae, is one of the most destructive diseases of the honeybee, Apis mellifera. Our group recently published the sequences of 9 new phages with the ability to infect and lyse P. larvae. Here, we characterize the genomes of these P. larvae phages, compare them to each other and to other sequenced P. larvae phages, and putatively identify protein function. The phage genomes are 38–45 kb in size and contain 68–86 genes, most of which appear to be unique to P. larvae phages. We classify P. larvae phages into 2 main clusters and one singleton based on nucleotide sequence identity. Three of the new phages show sequence similarity to other sequenced P. larvae phages, while the remaining 6 do not. We identified functions for roughly half of the P. larvae phage proteins, including structural, assembly, host lysis, DNA replication/metabolism, regulatory, and host-related functions. Structural and assembly proteins are highly conserved among our phages and are located at the start of the genome. DNA replication/metabolism, regulatory, and host-related proteins are located in the middle and end of the genome, and are not conserved, with many of these genes found in some of our phages but not others. All nine phages code for a conserved N-acetylmuramoyl-L-alanine amidase. Comparative analysis showed the phages use the “cohesive ends with 3′ overhang” DNA packaging strategy. This work is the first in-depth study of P. larvae phage genomics, and serves as a marker for future work in this area.
KEYWORDS: American Foulbrood, bacteriophage, comparative genomics, comparative proteomics, DNA packaging strategy, endolysin, large terminase, N-acetylmuramoyl-L-alanine amidase, Paenibacillus larvae, Phamerator, siphoviridae
Introduction
Paenibacillus larvae is a Gram-positive, spore-forming bacterium that is the causative agent of American Foulbrood Disease (AFB), one of the leading causes of the global population decline of the honeybee (Apis mellifera).1 As its name implies, P. larvae only infect the larva of the honeybee, adult bees being immune.2 Infection typically occurs when food contaminated with P. larvae spores is fed to a honeybee larva by nurse bees.3 The spores germinate and proliferate in the larval mid-gut within hours of ingestion, resulting in the death of the larva.2 The dead larvae turn into a viscous, brownish liquid that then dries to form a hard scale.3 AFB scales contain millions of highly infectious spores that are then inadvertently spread throughout the hive by other bees as they remove dead larvae from the hive.2 P. larvae spores are extremely durable, lasting several decades, and are largely antibiotic resistant, making treatment of P. larvae outbreaks difficult.3 Currently the only method for eliminating P. larvae outbreaks is the wholesale incineration of infected hives.2
Antibiotics such as oxytetracycline have been used extensively in the past to control AFB, however there now exist antibiotic-resistant P. larvae strains,4,5 and furthermore many countries ban the use of antibiotics on honeybees.2 As bees lack an adaptive immune system, one potential antibiotic-free AFB treatment is the use of bacteriophages that target P. larvae. Phages have several attractive features as a treatment strategy, such as not harming important symbiotic bacteria in the larval gut.6-9 The first P. larvae phages were identified from the 1950s through the 1990s, but these were not sequenced as rapid and cost-effective genome sequencing was not available at the time.10-18 With the advent of next-generation sequencing and the rise in antibiotic resistant P. larvae strains, there is growing interest in P. larvae phages as a potential treatment for AFB. In the last year alone 5 studies were published on treating AFB with P. larvae phages or P. larvae phage endolysins, with promising, if not conclusive, results.19-23
Since 2013, several bacteriophages that infect P. larvae were purified, sequenced, and characterized.23-26 Phage phiIBB_Pl23, isolated in Portugal in 2013, was the first to be sequenced and characterized,24 followed in 2015 by phages Diva, Lily, Rani, Redbud, Shelly and Sitara, isolated in North Carolina,25 and phage HB10c2 in Germany.23 Our group recently sequenced and published the genomes of 9 P. larvae phages.26
In this work, we characterize the genomes of these 9 new P. larvae phages and compare them to the genomes of other currently sequenced P. larvae phages. We putatively identify protein function and characterize the degree to which P. larvae phage proteins are conserved, with a focus on 2 phage proteins in particular: the large terminase and the N-acetylmuramoyl-L-alanine amidase endolysin.
Results
Phage sources, geographical origin, and morphology
The source and geographic origins of the 9 new phages are listed in Table 1. While 2 phages (Diane, Fern) were obtained from lysogens, all 9 phages lyse P. larvae in laboratory conditions (especially P. larvae genotype ERIC I) without needing to be induced, while leaving other Paenibacillus species unharmed.20 Electron micrographs of phages Diane (Fig 1A), Fern (Fig 1B) and Hayley (Fig 1C) are shown in Fig. 1. All of our phages are Siphoviridae, as are all currently known P. larvae phages.20,23-25 Capsids are prolate, approximately 100 nm long by 50 nm wide, and tails are approximately 150–200 nm long (Fig. 1).
Table 1.
Geographical origin and isolation source of P. larvae phages.
| Phage name | Geographical location | Isolation Source |
|---|---|---|
| Dianea | OH | Infected larva, ATCC culture 25747 |
| Fernb | USDA lab Germantown MD | Infected larva, P. larvae wild strain 2231 |
| Harrison | Gilcrease Orchards, N. Las Vegas, NV | Soil |
| Hayley | Gilcrease Orchards, N. Las Vegas, NV | Soil |
| Paisley | PA | Soil |
| Vadimc | NV | Lip balm |
| Vegasc | NV | Lip balm |
| Willow | Near Bremerton, WA | Soil |
| Xenia | USDA lab Germantown MD | Infected larva |
Lysogenic phage from ATCC culture 25747 isolated in Ohio by White from an infected insect27
Lysogenic phage from P. larvae wild strain 2231 isolated from an infected larva scale
Isolated from commercial products purchased in NV
Figure 1.
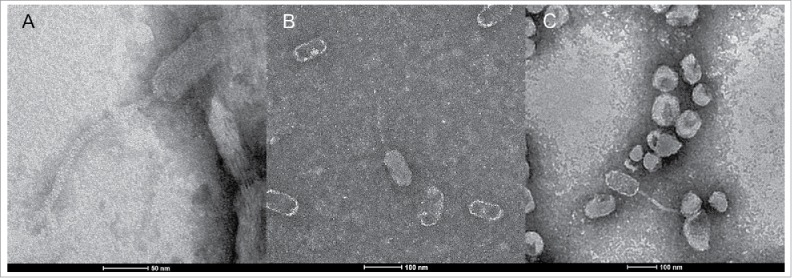
Scanning electron micrographs of phages (A) Diane, (B) Fern, and (C) Hayley.
Phage genome sequencing and assembly
The GenBank accession numbers and results of the genome assembly process for the 9 phages are shown in Table 2. Genome size ranges from 38 to 45 kb, and GC content from 40% to 43%. The genomes are 93–95% coding. No tRNAs were identified. The assembly process for Fern, Harrison, Paisley, Willow and Xenia produced complete genomes (hence min. coverage depth >1 ). For Diane, Hayley, Vadim and Vegas, the assembly process missed the genome ends (hence min. coverage depth = 1), and the genome ends were obtained by PCR as described in ref 26.
Table 2.
Accession numbers and genome assembly results of our P. larvae phages.
| Phage name | GenBank accession number | Genome length | Av. coverage depth | Min. coverage depth | GC content | Percent coding |
|---|---|---|---|---|---|---|
| Diane | KT361657 | 45,653 | 67 | 1 | 43.7 | 95.7 |
| Fern | KT361649 | 37,995 | 502 | 98 | 41.9 | 93.7 |
| Harrison | KT361651 | 44,247 | 291 | 61 | 40.2 | 93.6 |
| Hayley | KT361655 | 44,256 | 43 | 1 | 43.5 | 95.4 |
| Paisley | KT361653 | 44,172 | 350 | 58 | 40.0 | 93.5 |
| Vadim | KT361656 | 45,653 | 94 | 1 | 43.7 | 95.7 |
| Vegas | KT361654 | 45,653 | 128 | 1 | 43.7 | 95.7 |
| Willow | KT361650 | 37,994 | 122 | 50 | 41.9 | 93.7 |
| Xenia | KT361652 | 41,149 | 123 | 41 | 41.5 | 93.2 |
Genome annotation and comparative genomics of P. larvae phages
Genome annotation results are shown in Table 3. The genome annotation process identified between 68 and 86 protein coding genes in each phage. The number of genes increases linearly with genome size (R2=0.99). Approximately 90–95% of the P. larvae phage genes have a statistically significant BLASTP or CD-Search match (E-value<1E-3), while approximately half have a statistically significant BLASTP or CD-Search match to a protein with known function. Comparative genomics using our Phamerator database revealed that the majority (∼75%) of the genes are found only in P. larvae phages, with the majority of the remainder mostly shared with other Bacillus phages. Xenia has 9 genes not found in any other phages, while Harrison and Paisley each have one gene unique to them. Quantitative metrics of our phages genomes such as the length and number of non-coding gaps and overlaps are shown in Supplementary Table 1.
Table 3.
Comparative genomics of our P. larvae phages.
| Phage name | No. of genes | No. of genes with BLAST E-value <0.001 | No. of genes with putative function | Genes found in non-P. larvae phages | Genes found only in P. larvae phages | Gene unique to this phage |
|---|---|---|---|---|---|---|
| Diane | 86 | 83 | 45 | 22 | 64 | 0 |
| Fern | 68 | 65 | 36 | 18 | 50 | 0 |
| Harrison | 84 | 75 | 38 | 23 | 60 | 1 |
| Hayley | 84 | 81 | 43 | 21 | 63 | 0 |
| Paisley | 84 | 75 | 38 | 23 | 60 | 1 |
| Vadim | 86 | 83 | 45 | 22 | 64 | 0 |
| Vegas | 86 | 83 | 45 | 22 | 64 | 0 |
| Willow | 68 | 65 | 36 | 18 | 50 | 0 |
| Xenia | 77 | 72 | 43 | 20 | 48 | 9 |
Dotplots of the phages' genomes are shown in Fig. 2. All phages have a conserved region located at the start of the genome. Diane, Vadim, Vegas, and Hayley all appear to be highly similar to each other. Hayley appears to be missing a region located approximately in the middle of the genome that is present in Diane, Vadim, and Vegas. Paisley and Harrison are also very similar to each other, and Fern and Willow to each other. Xenia does not appear to be highly similar to any other phage, but seems closest to Fern and Willow.
Figure 2.
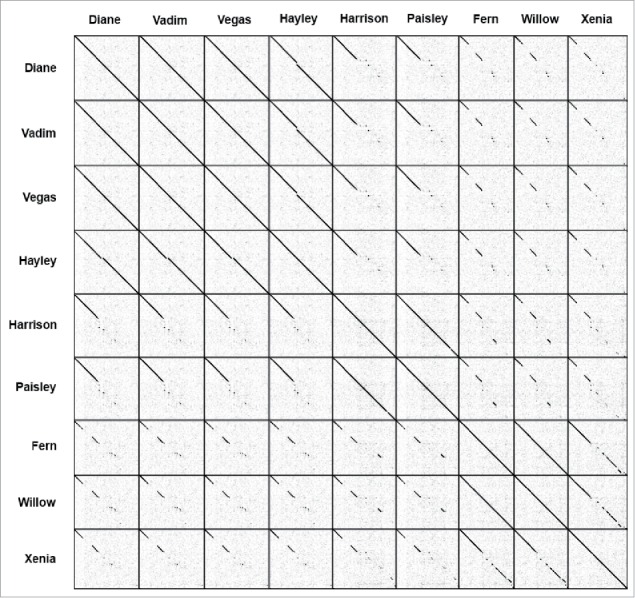
Dotplot of the genomes for 9 new P. larvae phages. A black dot is placed where there is nucleotide identity between 2 phages.
To quantify the degree of nucleotide sequence identity between our phages, we constructed a nucleotide sequence identity matrix using ClustalW, shown in Fig. 3. In this figure, we also included all other currently published P. larvae phages (Diva, Lily, Rani, Redbud, Shelly, Sitara, HB10c2 and phiIBB_Pl23). P. larvae phages fall into two similarity clusters containing phages with >60 % nucleotide sequence identity. Phage Lily is very divergent from all other P. larvae phages and does not fall into either cluster. Cluster A and Cluster B phages have a low degree of nucleotide sequence identity with each other (∼40%, which is roughly the percentage nucleotide sequence identity produced by ClustalW for 2 randomly generated nucleotide sequences of equal length). The clusters can be broken down into subclusters containing phages with >90 % nucleotide sequence identity, with Cluster B containing several singletons. All of these groupings cross geographical and source boundaries, e.g. Xenia (isolated in MD) has a very high degree of nucleotide sequence identity (99.5%) with Shelly (isolated in NC). Phages within the same subcluster have similar, though not identical lytic profiles and plaque morphologies.20 For this reason, we considered Diane, Vadim and Vegas (>99.9% nucleotide sequence identity) and Fern and Willow (99.99% nucleotide sequence identity) to be distinct from each other. However, as the annotation process did not produce any differences between Diane, Vadim and Vegas, we treat these phages as one in subsequent genomic analyses, and do likewise for Fern and Willow. In contrast, annotations of Xenia and Shelly are not identical despite the 99.5% nucleotide sequence identity between these two phages, as Shelly was annotated by another group.
Figure 3.
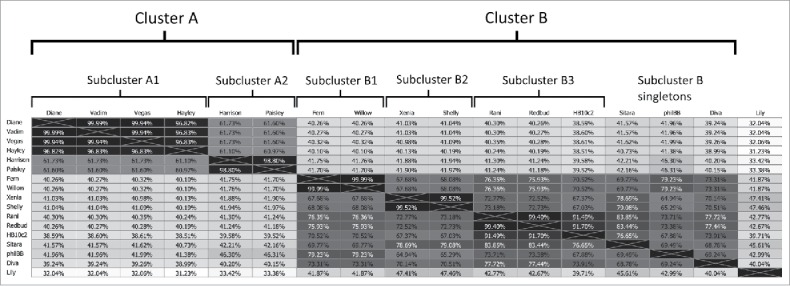
Percent nucleotide sequence identity matrix for all 17 sequenced P. larvae phages. Phages are classified into clusters and subclusters based on nucleotide sequence identity.
Genome maps produced with Phamerator are shown in Figs. 4 and 5. The difference between Diane/Vadim/Vegas and Hayley is due to 2 genes absent in Hayley (gp30 and gp31), but otherwise all 4 of these phages contain the same genes. Similarly, the difference between Harrison and Paisley is due to a single gene (gp65). Up to gp21 the majority of genes appear to be fully conserved in all phage genomes, with the exception of gp5 and gp8–15, which differ between Cluster A phages and Fern/Willow and Xenia. Of these, gp5, gp8, gp14 and gp15 are single pham genes and are thus still somewhat conserved across all phages, however gp9–13 are not. Past gp21 the genomes diverge, the sole exception being Xenia and Fern/Willow, which have several genes in the same pham throughout the genome, especially in the region between gp42 and gp52 in Fern/Willow (gp51 and gp61 in Xenia). Gp53 and gp54 in Harrison and Paisley are very similar to gp40 and gp41 in Fern/Willow, even though they occur in a non-conserved region in the genomes' mid-section, possibly indicating horizontal gene transfer. The same is true of the last gene in the genome of all the phages.
Figure 4.
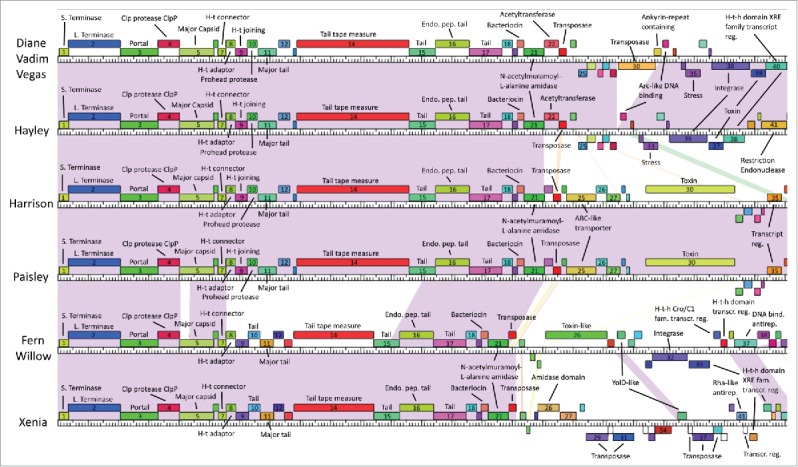
Genome maps of our phages obtained from Phamerator (first half). Boxes represent genes, with boxes of the same color indicating genes in the same pham. Genes in a pham of their own are uncolored. Shaded areas indicate regions of high nucleotide sequence similarity between phages, with purple indicating the highest degree of similarity, and red the lowest.
Figure 5.
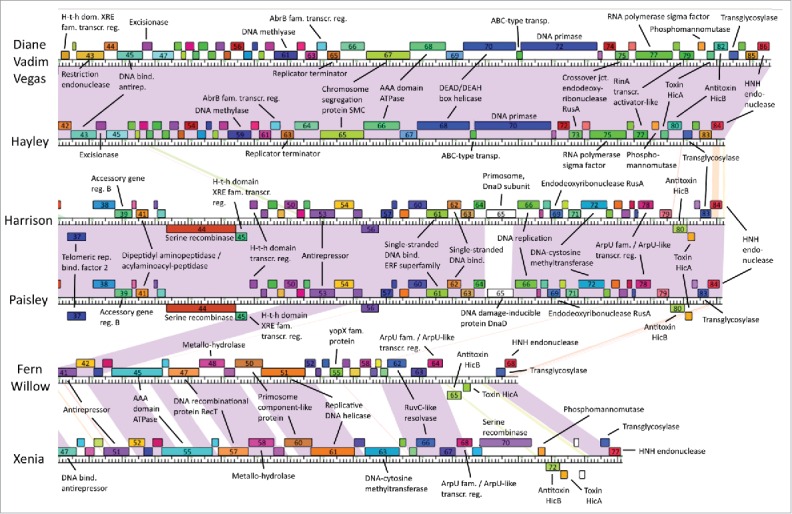
Genome maps of our P. larvae phages obtained from Phamerator (second half). Boxes represent genes, with boxes of the same color indicating genes in the same pham. Genes in a pham of their own are uncolored. Shaded areas indicate regions of high nucleotide sequence similarity between phages, with purple indicating the highest degree of similarity, and red the lowest.
P. larvae phage protein functions
Gene products that have at least one statistically significant (E-value <1E-3) BLAST or CD-Search match with a protein of known function are shown in Table 4. The list of all the gene products of our 9 P. larvae phages, the phams to which they belong, and any other phage gene products in those phams are included as Supplementary Table 2.
Table 4.
P. larvae phage genes with statistically significant BLAST and/or CDD matches (E-value < 1E-3) to proteins with known function. The gene product number is shown in the first row of each cell, and the pham number is shown in the second row, italicized in parentheses. Rows are colored according to protein function. We classify phage proteins into 6 functional categories: 1) virion particle (blue), 2) virion assembly (burgundy), 3) host lysis (purple), 4) DNA replication/metabolism (tan), 5) gene regulation, including putative transcription factors (green), and 6) host-related functions (yellow). Gene products whose function cannot be classified into these 6 categories due to lack of sufficient information or conflicting information are left uncolored. Instances where there are 2 or more unrelated functions with statistically significant matches are marked with a footnote, with the more statistically significant (or with higher bit score) function listed in the table, and the less statistically significant (or with lower bitscore) function listed in the footnotes at the end of the table.
| Diane/Vadim/Vegas | Hayley | Harrison | Paisley | Fern/Willow | Xenia | |
|---|---|---|---|---|---|---|
| Small terminase | gp1 (16596) | gp1 (16596) | gp1 (16596) | gp1 (16596) | gp1 (16596) | gp1 (16596) |
| Large terminase | gp2 (16567) | gp2 (16567) | gp2 (16567) | gp2 (16567) | gp2 (16567) | gp2 (16567) |
| Portal protein | gp3 (16597) | gp3 (16597) | gp3 (16597) | gp3 (16597) | gp3 (16597) | gp3 (16597) |
| Clp protease ClpP | gp4 (16598) | gp4 (16598) | gp4 (16598) | gp4 (16598) | gp4 (16598) | gp4 (16598) |
| Major capsid protein | gp5 (16599) | gp5 (16599) | gp5 (16599) | gp5 (16599) | gp5 (16599) | gp5 (16599) |
| Head-tail connector protein | gp7a(16601) | gp7a(16601) | gp7a(16601) | gp7a(16601) | gp7a(16601) | gp7a(16601) |
| Head-tail adaptor protein | gp8b(16602) | gp8b(16602) | gp8b(16602) | gp8b(16602) | gp8b(16602) | gp8b(16602) |
| Head-tail joining protein | gp9 (16865) | gp9 (16865) | gp9 (16865) | gp9 (16865) | ||
| Prohead protease | gp10 (16866) | gp10 (16866) | gp10 (16866) | gp10 (16866) | ||
| Tail protein | gp10 (16604) | gp10 (16604) | ||||
| Major tail protein | gp11 (16867) | gp11 (16867) | gp11 (16867) | gp11 (16867) | gp11 (16605) | gp11 (16605) |
| Tail tape measure protein | gp14 (16578) | gp14 (16578) | gp14 (16578) | gp14 (16578) | gp14 (16578) | gp14 (16578) |
| Tail protein | gp15 (16608) | gp15 (16608) | gp15 (16608) | gp15 (16608) | gp15 (16608) | gp15 (16608) |
| Endopeptidase tail protein | gp16c(16609) | gp16c(16609) | gp16c(16609) | gp16c(16609) | gp16c(16609) | gp16c (16609) |
| Tail protein | gp17 (16610) | gp17 (16610) | gp17 (16610) | gp17 (16610) | gp17 (16610) | gp17 (16610) |
| Bacteriocin biosynthesis protein | gp20d(16613) | gp20d(16613) | gp20d(16613) | gp20d(16613) | gp20d(16613) | gp20d(16613) |
| N-acetylmuramoyl-L-alanine amidase | gp21 (16614) | gp21 (16614) | gp21 (16614) | gp21 (16614) | gp21 (16614) | gp21 (16614) |
| Acetyltransferase | gp22e(16870) | gp22e(16870) | ||||
| Transposase | gp23f(16615) | gp23f(16615) | gp23f(16615) | gp23f(16615) | gp22f(16615) | gp22f(16615) |
| ABC-like transporter protein | gp25 (19147) | gp25 (19147) | ||||
| Amidase domain protein | gp26g(16618) | |||||
| Transposase | gp30 (16876) | gp29h(16621) | ||||
| Transposase | gp31 (16622) | |||||
| Toxin-like protein | gp26 (18320) | |||||
| YolD-like protein | gp28 (16625) | gp35 (16625) | ||||
| Toxin | gp30 (19152) | gp30 (19152) | ||||
| Ankyrin-repeat containing protein | gp31i(16877) | |||||
| Arc-like DNA binding protein | gp32 (16878) | gp30 (16878) | ||||
| Transposase | gp36h(690) | |||||
| Transposase | gp37h(16626) | |||||
| Transposase | gp38 (16627) | |||||
| Stress protein | gp35 (16880) | gp33 (16880) | ||||
| Integrase | gp38 (16588) | gp36 (16588) | gp32 (16588) | |||
| Toxin | gp39 (16723) | gp37 (16723) | ||||
| Transcriptional regulator | gp35 (19155) | gp35 (19155) | ||||
| Telomeric repeat binding factor 2 | gp37 (19014) | gp37 (19014) | ||||
| Accessory gene regulator B | gp39 (16970) | gp39 (16970) | ||||
| Dipeptidyl aminopeptidase/ acylaminoacyl-peptidase | gp41 (19158) | gp41 (19158) | ||||
| Serine recombinase | gp44j(19161) | gp44j(19161) | ||||
| Helix-turn-helix domain XRE family transcriptional regulator | gp40 (16883) | gp38 (16883) | ||||
| Helix-turn-helix domain XRE family transcriptional regulator | gp41 (16884) | gp39 (16884) | gp45 (19162) | gp45 (19162) | gp33k(17376) | |
| Helix-turn-helix Cro/C1 family transcriptional regulator | gp34 (18325) | |||||
| Helix-turn-helix domain transcriptional regulator | gp35l(18326) | |||||
| Helix-turn-helix domain XRE family transcriptional regulator | gp46 (19163) | gp46 (19163) | ||||
| Rha-like antirepressor | gp41 (16810) | |||||
| Transcriptional regulator | gp42m(696) | |||||
| Helix-turn-helix domain XRE family transcriptional regulator | gp43n(16628) | |||||
| Helix-turn-helix domain XRE family transcriptional regulator | gp44 (16629) | |||||
| Restriction endonuclease | gp43 (16885) | gp41 (16885) | ||||
| DNA binding antirepressor | gp45o(16631) | gp43o(16631) | gp37o(16631) | gp47o(16631) | ||
| Excisionase | gp46 (16887) | gp44 (16887) | ||||
| Antirepressor | gp53 (16689) | gp53 (16689) | gp41 (16689) | gp51 (16689) | ||
| AAA domain ATPase | gp45 (16634) | gp55 (16634) | ||||
| DNA recombinational protein RecT | gp47 (16636) | gp57 (16636) | ||||
| Metallo-hydrolase | gp48 (16637) | gp58 (16637) | ||||
| Primosome component-like protein | gp50 (16639) | gp60 (16639) | ||||
| Replicative DNA helicase | gp51 (16640) | gp61 (16640) | ||||
| yopX family protein | gp55 (16645) | |||||
| DNA methylase | gp61 (16902) | gp59 (16902) | ||||
| Single-stranded DNA binding protein, ERF superfamily | gp61 (19169) | gp61 (19169) | ||||
| Single-stranded DNA binding protein | gp62 (16720) | gp62 (16729) | ||||
| AbrB family transcriptional regulator | gp64 (16905) | gp62 (16905) | ||||
| Replication terminator protein | gp65 (16906) | gp63 (16906) | ||||
| Primosome, DnaD subunit | gp65 (16529) | |||||
| DNA damage-inducible protein DnaD | gp65 (15038) | |||||
| DNA replication protein | gp66p(16552) | gp66p(16552) | ||||
| Chromosome segregation protein SMC | gp67q(16908) | gp65q(16908) | ||||
| AAA domain ATPase | gp68r(16909) | gp66r(16909) | ||||
| DEAD/DEAH box helicase | gp70 (16911) | gp68 (16911) | ||||
| ABC-type transport system, ATP binding protein | gp71 (16912) | gp69 (16912) | ||||
| DNA primase | gp72s(16913) | gp70s(16913) | ||||
| Endodeoxyribonuclease RusA | gp69 (19173) | gp69 (19173) | ||||
| Crossover junction endodeoxyribonuclease RusA | gp75 (16916) | gp73 (16916) | ||||
| DNA–cytosine methyltransferase | gp72 (16642) | gp72 (16642) | gp63 (16642) | |||
| RuvC-like resolvase | gp62 (16647) | gp66 (16647) | ||||
| RNA polymerase sigma factor | gp77 (16918) | gp75 (16918) | ||||
| ArpU family/ArpU-like transcriptional regulator | gp78 (16560) | gp78 (16560) | gp64 (16560) | gp68 (16560) | ||
| RinA transcriptional activator-like protein | gp79 (16920) | gp77 (16920) | ||||
| Serine recombinase | gp70 (16818) | |||||
| Phosphomannomutase | gp80 (16819) | gp78 (16819) | gp71 (16819) | |||
| Toxin HicA | gp81t(16921) | gp79t(16921) | ||||
| Antitoxin HicB | gp82 (16922) | gp80 (16922) | gp80 (16649) | gp80 (16649) | gp65 (16649) | gp72 (16649) |
| Toxin HicA | gp81t(16820) | gp81t(16820) | gp66 (16650) | gp73t(16820) | ||
| Transglycosylase | gp83 (16651) | gp81 (16651) | gp83 (16651) | gp83 (16651) | gp67 (16651) | gp 76 (16651) |
| HNH endonuclease | gp86 (16652) | gp84 (16652) | gp84 (16652) | gp84 (16652) | gp68 (16652) | gp77 (16652) |
Also has equally strong BLAST and CDD matches to DNA packaging protein
Also has equally strong BLAST and CDD matches to head-tail joining protein
CDD matches only (Evalue=1E-123)
Also has strong BLAST and CDD matches to bhlA protein
Also has equally strong BLAST matches to DNA methyltransferase
Also has strong BLAST matches to holin
Also has equally strong BLAST matches to peptidase domain
Also has equally strong BLAST and CDD matches to integrase
Also has strong BLAST matches to toxin-like protein, DNA Smf single strand binding protein, transcriptional regulatory protein YclJ, phosphatase, transposase
Also has equally strong BLAST matches to integrase, ATPase, resolvase, invertase
Also has strong BLAST and CDD matches to peptidase
Also has strong BLAST and CDD matches to excisionase
Also has strong BLAST matches to Xre-like protein
Also has strong BLAST matches to repressor
Also has equally strong BLAST and CDD matches to Rha family transcriptional regulator
Also has equally strong BLAST and CDD matches to chromosomal replication initiator protein DnaA
Also has equally strong BLAST matches to DNA recombination protein RecF
Also has strong BLAST matches to oxidoreductase, putative DNA helicase, putative RecA NTPase, ATP-dependent Lon protease
Also has equally strong BLAST matches to RecA familyATPase
Also has equally strong BLAST and CDD matches to ycfA-like protein
Virion particle genes
Virion particle genes are clustered near the start of the genome, from position gp3 to gp17. They include a portal protein (gp3), a major capsid protein (gp5), a head-tail connector (gp7), a head-tail adaptor protein (gp8) and a head-tail joining protein (gp9), and 5 or 6 tail proteins, including a major tail protein (gp11), a tail tape measure protein (gp14), and an endopeptidase tail protein (gp16). The head-tail adaptor protein at gp7 also has strong BLAST and CD-Search matches with a “DNA packaging protein.” However, as this is not confirmed and DNA packaging is handled by the terminase, we assigned head-tail adaptor function to this gene product. The tail tape measure protein is encoded by the longest gene in the genome in all of the phages. All the identified virion particle genes are found in all of our phages, except for the head-tail joining protein (gp9) found only in Cluster A phages, and a tail protein (gp10) exclusive to Fern/Willow and Xenia.
Virion particle genes are conserved, as they are all single-pham genes, the sole exception being the major tail protein (gp11), which is in 2 phams (see also Fig. 3); one pham for the Cluster A phages (who have the same major tail protein), and one pham for Fern/Willow, and Xenia (who also have the same major tail protein). The tail tape measure protein (gp14) is a single-pham gene, however it follows the same pattern as the major tail protein: Cluster A phages have an identical tail tape measure protein that is different than the tail tape measure protein of Fern/Willow, and Xenia. Both the major tail protein and tail tape measure protein of Cluster A phages are considerably longer than those of Fern/Willow and Xenia, suggesting Cluster A phages have longer tails than Fern/Willow and Xenia. From Fig. 1, we can discern that Diane and Hayley do indeed have a longer tail (∼200 nm) than Fern (∼150 nm).
Virion assembly genes
Assembly genes identified include a small and large terminase (gp1 and gp2, respectively), a Clp protease (gp4) and a prohead protease (gp10). The small and large terminase and the Clp protease are found in all the phages, but the prohead protease at gp10 is only present in Cluster A phages. The assembly genes are all conserved, all of them being single-pham genes.
Host lysis genes
All of our phages encode an N-acetylmuramoyl-L-alanine amidase endolysin at position gp21. This protein varies between 224 and 226 amino acids in length and is conserved among our phages, as all N-acetylmuramoyl-L-alanine amidases are in a single pham. In addition, all of our phages encode a transglycosylase near the end of their genomes. Transglycosylases, also known as glycosyltransferases, are known to cleave glycosidic bonds in the host glycan, and are thus used by phages for host lysis.28-30 The transglycosylase is conserved, as it is a single-pham gene. In addition, Xenia encodes a protein (gp26) with statistically significant matches to an amidase domain, although nothing more is known about the function of this protein (and it also has statistically significant matches to peptidase domains).
DNA replication and metabolism genes
All our phages encode numerous genes with putative functions related to DNA replication and metabolism. These include transposases, integrases, endonucleases, serine recombinases, excisionases, methyltransferases, and others. This is by far the largest and most diverse functional category. The vast majority of DNA replication and metabolism genes are not conserved among our phages. Only 2 genes in this category, the transposase at gp23/gp22 and the HNH endonuclease (which is the last gene in the phage genomes), are found in all the phages and are conserved. Of significance is that the transposase at gp23/gp22 has significant BLAST matches to proteins with holin function. However, the matches to transposase function are much more statistically significant than to those with holin function (e.g., E-value of 1E-37 compared to 1E-5), thus we assigned it transposase function.
A conserved integrase is found in Diane/Vadim/Vegas (gp38), Hayley (gp36), and Fern/Willow (gp32). Harrison and Paisley possess a serine recombinase at gp44 that has equally significant BLAST and CDD matches to integrases, therefore this protein could be an integrase. In addition, Xenia possesses several genes assigned transposase function that have equally significant matches to proteins with integrase function (gp29, gp36, gp37). It is therefore possible and in fact likely that all of our phages possess at least one integrase, indicating they possess lysogenic potential; in fact 2 of our phages (Diane and Fern) were isolated as lysogens that converted to lytic phages in vitro.
Regulatory genes
All 9 P. larvae phages encode genes that regulate gene expression, whether in the host or the phage itself. These include XRE (Xenobiotic response element), Cro/Cl, AbrB (ambiactive repressor) and ArpU (autolysin regulatory protein) family transcriptional regulators, as well as anti-repressor proteins. Many of these proteins, in particular the XRE-family transcriptional regulators, contain a helix-turn-helix domain. However, the function of these proteins in the P. larvae phage life cycle is not known, which suggests these phages use novel methods of gene regulation to modulate host expression in support of their life cycle. These are the least conserved genes in our phages. There is no regulatory gene that is common to all of the phages, and even genes of the same family are divergent, e.g., the XRE family transcriptional regulator at gp41 in Diane/Vadim/Vegas, gp39 in Hayley, gp45 in Harrison and Paisley, and gp33 in Fern/Willow, is in 3 different phams.
Host-related genes
The nine phages also code for a variety of host-related proteins, such as several toxins, 2 ABC transporters, a stress protein, a metallo-hydrolase, a phosphomannomutase, a toxin-antitoxin system, and others. At position gp20 all the phages code for a conserved (single-pham) bacteriocin, a toxin prokaryotes produce to inhibit the growth of closely related competitor strains.31 This gene also has strong BLAST matches to a “bhlA protein,” an unconfirmed holin-like protein.32,33 While this could be the “missing” holin gene, we assigned bacteriocin function due to its much more statistically significant match (E-value < 1E-100 compared to 1E-13). Fern/Willow and Xenia contain a putative metallo-hydrolase, a type of β-lactamase (gp48/58). All of the phages also encode the HicA/HicB toxin/antitoxin system. With the exception of the bacteriocin, none of these genes are conserved. Besides the bacteriocin, only the toxin-antitoxin genes are present in all of the phages, and these are not conserved; the HicA genes are in 3 phams, while the HicB genes are in 2 phams. In Diane/Vadim/Vegas and Hayley the HicA genes are located in front of the HicB genes, while the opposite is true in the other phages.
Gene operons
In every one of our phages' genomes, there are 10 to 15 instances of genes whose start codon is located 3 bp before the stop codon of the gene upstream, suggesting these genes are transcribed together as part of an operon. Of these, the following are operons involving proteins with putative function: The large terminase at gp2 and the portal protein at gp3 (all phages), the Clp protease at gp4 and the major capsid protein at gp5 (all phages), the head-tail connector at gp7 and the head-tail adaptor at gp8 (all phages), which extends to include the head-tail joining protein at gp9 and the prohead protease at gp10 in the Cluster A phages, the major tail protein at gp11 and the hypothetical protein at gp12 (Fern/Willow, Xenia), the tail tape measure protein at gp14 and the tail protein at gp15 (Fern/Willow, Xenia), the endopeptidase tail protein at gp16 and the tail protein at gp17 (all phages), the bacteriocin at gp20 and the N-acetylmuramoyl-L-alanine amidase at gp21 (all phages), the transglycosylase at gp67/gp76 and the HNH endonuclease at gp68/gp77 (Fern/Willow, Xenia).
Multiple alignment of P. larvae phage large terminases
We performed a multiple alignment of the P. larvae phage large terminases using ClustalW in Fig. 6. The alignment showed that there are only 3 distinct large terminases for the 17 known P. larvae phages. Diane/Vadim/Vegas, Hayley, Harrison and Paisley all have the same large terminase (Group 1), as do Fern/Willow, Xenia, Diva, Rani, Redbud, Shelly, Sitara, HB10c2 and phiIBB_Pl23 (Group 2), with the large terminase of Lily by itself (Group 3). The large terminases follow the classification of the phages based on nucleotide sequence identity, i.e. Cluster A phages all have the Group1 large terminase, while Cluster B phages have the Group 2 large terminase, with Lily an outlier (Fig. 3). From Fig. 6A we observe 11 locations where the Group 1 and Group 2 large terminases differ, corresponding to an amino acid sequence identity of approximately 98% (Fig. 6B). We also note the large terminase of Lily is very distant from the other 2 (10% amino acid identity), and also considerably longer than the other 2 (622 amino acids compared to 574 amino acids). A pham circle of the P. larvae phage large terminase is included as Supplementary Fig. 1.
Figure 6.
Multiple alignment and percent amino acid sequence identity matrix of P. larvae phage large terminases. With the exception of Lily, all P. larvae phages have a large terminase that is either identical to that of Diane (Diane, Hayley, Vadim, Vegas, Harrison, Paisley), or Xenia (Xenia, Fern, Willow, Diva, Rani, Redbud, Shelly, Sitara, HB10c2, phiIBB_Pl23).
Multiple alignment of P. larvae phage N-acetylmuramoyl-L-alanine amidases
A multiple alignment of the N-acetylmuramoyl-L-alanine amidases of all the P. larvae phages is shown in Fig. 7. There are 5 distinct N-acetylmuramoyl-L-alanine amidases among the 17 currently sequenced P. larvae phages. Group 1 consists of the N-acetylmuramoyl-L-alanine amidase of Diane/Vadim/Vegas, and Hayley, Group 2 consists of Harrison, Paisley, and phiIBB_Pl23, Group 3 consists of Fern/Willow, Group 4 consists of Xenia, Diva, Shelly and Sitara, and Group 5 consists of HB10c2, Redbud, and Rani. The N-acetylmuramoyl-L-alanine amidases follow the classification of the phages based on nucleotide sequence identity (Fig. 3), i.e. phages in the same subcluster have the same N-acetylmuramoyl-L-alanine amidase. The 5 different N-acetylmuramoyl-L-alanine amidases all have >90 % similarity with each other. A pham circle of the P. larvae phage N-acetylmuramoyl-L-alanine amidases is included as Supplementary Fig. 2.
Figure 7.
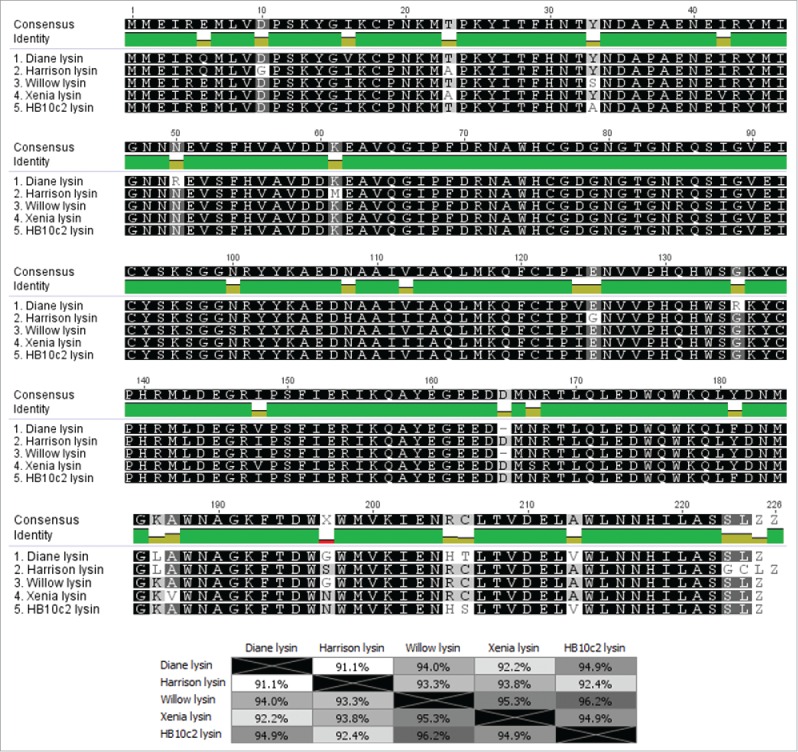
Multiple alignment and average amino acid identity matrix of P. larvae phage N-acetylmuramoyl-L-alanine amidases. There are 5 distinct P. larvae phage N-acetylmuramoyl-L-alanine amidases, with phages in the same group having an identical N-acetylmuramoyl-L-alanine amidase. Group 1 consists of phages Diane, Vadim, Vegas and Hayley, Group 2 consists of phages Harrison, Paisley and phiIBB_Pl23, Group 3 consists of phages Willow and Fern, Group 4 consists of phages Xenia, Shelly, Diva, and Sitara, and Group 5 consists of phages HB10c2, Rani, and Redbud.
P. larvae phages use the 3′ cohesive ends DNA packaging strategy
Comparative analysis shows our phages use the cohesive ends with 3′ overhangs DNA packaging strategy. Diva, Rani, Redbud, Shelly and Sitara (whose large terminases are either 98% or 100% identical with those of our phages) are known to possess 9-bp 3′ overhangs with the sequence “CGACTGCCC.”25 We found the same 9-bp sequence in Diane/Vadim/Vegas, Hayley, Fern/Willow and Xenia, and rearranged their genomes so that base 1 is the first base immediately after the last overhang base. When the genomes are rearranged this way, the first gene in the genome is the small terminase, which begins 50 bp downstream of base 1, exactly like in Diva, Rani, Redbud, Shelly and Sitara.25 When we rearrange the genomes of Harrison and Paisley in this manner, this reveals an overhang whose sequence is “CGACGGACC,” differing by 2 bases from the overhang of the other phages, even though the large terminase of Harrison and Paisley is identical to that of Diane/Vadim/Vegas and Hayley. That P. larvae phages use the 3′ cohesive ends packaging strategy is further confirmed by a phylogenetic tree of the large terminases of the phages in our Phamerator database, shown in Supplementary Fig. 3.
Discussion
In this study we have conducted an in-depth comparative genomic analysis of 9 P. larvae phages recently sequenced and published by our group. These phages were isolated from a variety of sources, such as infected larvae, soil samples, and commercial beeswax products, from different geographical regions of the United States. Interestingly, there are several instances of phages from different locations having a very high degree of nucleotide sequence identity with each other. Phage Fern (lysogenic phage isolated from a wild P. larvae strain) is very similar to phage Willow (soil sample from Washington state); phage Harrison (soil sample from Nevada) is very similar to phage Paisley (soil sample from Pennsylvania); and phages Diane (lysogenic phage isolated from ATCC P. larvae strain), Vadim (commercial beeswax product), Vegas (another commercial beeswax product) and Hayley (soil sample from Nevada) are all very similar to one another. Phage Xenia (isolated from infected larva from a USDA lab in Maryland) shows a very high degree of sequence similarity (99.5%) with phage Shelly, which was isolated in North Carolina by another group. These findings suggest that subsets of P. larvae phages are subject to very similar selection pressures.
P. larvae phages can be classified into 2 main clusters based on nucleotide sequence identity (we used a threshold of 60%), both of which can be broken down into 2 or more subclusters, and one singleton (Lily). Cluster A phages (Diane, Vadim, Vegas, Hayley, Harrison, Paisley) show little sequence similarity (∼40%) with Cluster B phages (Fern, Willow, Xenia, Diva, Rani, Redbud, Shelly, Sitara, HB10c2, phiIBB_Pl23). The clusters are themselves heterogeneous and can be further broken down into subclusters that contain phages that are very similar to one another (>90% nucleotide sequence identity), and in the case of Cluster B, several singletons. This is similar to what has been observed in other well-studied phages, such as Mycobacterium phages.34 As with Mycobacterium phages, we expect that as the number of sequenced P. larvae phages increases over time, the clusters and subclusters will increase in number and grow in size and diversity.35
Comparative genomic analysis of the 9 new P. larvae phages shows that the majority of their genes are only found in P. larvae phages. Using bioinformatics tools alone, we were able to predict putative functions for about half of the genes of the new P. larvae phages. We found genes coding for virion particle proteins, virion assembly proteins, host lysis proteins, DNA replication and metabolism proteins, regulatory proteins, and host-related proteins. Almost all of the virion particle and assembly genes are found in all our phages and are conserved, indicating similar morphology and assembly mechanisms. The tail proteins may possess catalytic activity (e.g., gp16 may have endopeptidase activity), which would allow the phages to penetrate into their host; more work is needed to understand how P. larvae phages invade their hosts. On the other hand, the DNA replication/metabolism, regulatory genes, and host-related genes are generally not conserved. Many of the DNA replication/metabolism, regulatory, and host-related genes are found in some of the phages but are absent from others. This suggests diverse and potentially novel DNA replication and gene regulation mechanisms at the transcriptional level. More work is needed to understand the functions of many of the DNA replication/metabolism, regulatory, and host-related genes, as their precise role in P. larvae phage and/or P. larvae biology is not known. The host-related genes are of particular interest as they include genes implicated in antibiotic resistance, such as a β-lactamase, and host virulence, such as toxins, a bacteriocin, and a toxin-antitoxin system. These genes may be used by the phages, once integrated into the host chromosome, to promote their spread by assisting infected P. larvae in outcompeting bacterial competitors and in defending against antibiotics.
In terms of genome architecture, the conserved virion particle and assembly genes are located at the front end of the genome in synteny, typical of Siphoviridae phage genomes.36 It is possible, and in fact likely, that genes located in this genomic region whose function cannot be inferred from sequence comparison alone, such as gp6, gp9, gp12, gp13, gp18 and gp19 encode virion particle or assembly genes, but more work is needed to identify the function of these genes. The divergent DNA replication/metabolism, regulatory and host-related genes are located downstream of the virion particle and assembly genes. The genomes of our P. larvae phages converge at the ends, where a conserved transglycosylase and HNH endonuclease are located.
All of the new phages encode a highly conserved N-acetylmuramoyl-L-alanine amidase endolysin. Multiple alignment of the P. larvae phages' N-acetylmuramoyl-L-alanine amidase revealed that there are 5 distinct N-acetylmuramoyl-L-alanine amidases among the 17 currently sequenced P. larvae phages, all with >90% amino acid sequence identity to each other. Phages grouped in the same subcluster by nucleotide sequence identity have the same N-acetylmuramoyl-L-alanine amidase, suggesting subsets of P. larvae phages lyse slightly different hosts.
Many bacteriophages lyse their hosts by means of a holin/endolysin cassette.37 The new P. larvae phages seem to lack a holin on first inspection, although they do encode for at least 2 proteins with significant matches to holin or holin-like proteins (the bacteriocin at gp20 and the transposase at gp23/gp22). It could be that either (or perhaps both) of these proteins are indeed holins used by the P. larvae phages. This possibility is reinforced by the fact that both genes are located proximally to the N-acetylmuramoyl-L-alanine amidase, and that the putative bacteriocin is part of the same operon with the N-acetylmuramoyl-L-alanine amidase. Holins are generally not conserved, and are therefore difficult to detect bioinformatically, thus more work is needed in this area.
The phages also code for a transglycosylase, raising the interesting possibility that P. larvae phages have more than one lytic mechanism. The fact that the transglycosylase is found in all our phages and is conserved (all transglyosylases are in the same pham), lends additional support to this hypothesis. This gene occurs in a region of the genome that is not conserved, suggesting it may have spread by horizontal gene transfer. More work is needed to discern the mechanisms of how P. larvae phages lyse their hosts.
All the phages also encode at least one transposase and likely one integrase. Thus in addition to lytic activity, they also appear to possess lysogenic activity. However these proteins are not conserved among our phages, pointing to potentially different lysogenic mechanisms.
Analysis of the large terminase protein indicated that there are only 3 distinct large terminases among the 17 currently sequenced P. larvae phages. Two of the large terminases are very similar to each other, having 98% amino acid sequence identity between them. These 2 large terminases account for 16 of the 17 P. larvae phages, the sole exception being the large terminase of phage Lily, which is very divergent from the other two (i.e. Cluster A and Cluster B). Phages in the same cluster have the same large terminase. All our P. larvae phages use the “cohesive ends with 9-bp 3′ overhangs” strategy, consistent with all other sequenced P. larvae phages (with the sole exception of phage Lily).
In recent years there has been a surge of interest in P. larvae phages, partly due to their potential to treat AFB. The number of sequenced P. larvae phages has increased from 0 at the start of 2013, to 17 as of this writing, and is likely to grow significantly. Our comparative genomic study is the first of its kind, and we expect to see much growth in P. larvae genomics in the coming years. Key areas to be addressed are identifying the function of more P. larvae phage proteins, the evolutionary history of P. larvae phages, the mechanisms by which P. larvae phages lyse their hosts, including identification of P. larvae phage holins and the role of transglycosylase, and the role of phage-encoded β-lactamases and toxins in P. larvae antibiotic resistance and virulence. Other potential areas of interest are the mechanism by which P. larvae phages penetrate their host, the relationship of P. larvae phages to their hosts in the wild, including the phages' role in horizontal gene transfer, identifying uses of P. larvae phage proteins for biotechnology applications, understanding how P. larvae defend against infection from phages, and further studies on the use of P. larvae phages as a treatment of AFB.
Materials and methods
Phages were isolated from a variety of sources and amplified using P. larvae NRRL 2605. Details of the isolation and amplification process are given in20. Assembly was carried out using Geneious 7.1 (Biomatters, Auckland, NZ).38 Details of the assembly process are given in26.
Genomes were annotated using DNA Master (cobamide2.bio.pitt.edu), which includes the gene calling programs Glimmer (http://ccb.jhu.edu/software/glimmer)39 and GeneMark (exon.gatech.edu).40 We also used GeneMark.hmm (exon.gatech.edu).41 Details of the annotation procedure are given in26.
Dot plots were obtained with Gepard 1.30 (cube.univie.ac.at/gepard).42 The percent nucleotide sequence identity between phage genomes was obtained by performing a multiple alignment using ClustalW,43 using the IUB cost matrix. Protein alignments were performed using ClustalW using the BLOSUM62 cost matrix. Protein phylogenetic trees were constructed using Clustal Omega.44
Putative protein function was inferred from manual curation of searches of NCBI's non-redundant protein database with BLASTP, and searches of NCBI's Conserved Domain Database (CDD) with CD-Search,45 both with an E-value cutoff of 1E-3. In cases where the searches returned multiple conflicting results, the result with the lowest E-value was chosen (unless the result was a “hypothetical protein,” in which case the result with the lowest E-value that wasn't a hypothetical protein was entered). In cases where there were conflicting results with equal E-value, the bit score was used as a tie-breaker.
Phage genome maps and pham circles were obtained from Phamerator.46 Phage genome maps were obtained using the “Align Two Sequences” algorithm of BLASTN and default window and step size, and an E-value cutoff of 1E-4. Genes with percent nucleotide identity >32.5% as calculated using ClustalW and BLAST E-value < 1E-50 were grouped into the same “pham”. The Phamerator database was populated with Bacillus and non-Bacillus phages whose proteins appeared in our BLAST results with E-value < 1E-3, as in47. The full list of phages in our Phamerator database, their accession number, and host, is included as Supplementary Table 3.
Supplementary Material
Disclosure of potential conflicts of interest
Amy, PS and DG Yost disclose patent US 20140213144A1. Amy, PS and L LeBlanc disclose patent WO2015153956.
Acknowledgments
We would like to acknowledge Bryan Merrill and Sandra Burnett of BYU for help in setting up the Phamerator database and numerous helpful discussions during the genome annotation process. We would also like to thank all who contributed samples for phage isolation, as well as Andrew Krohn of Northern Arizona University for DNA sequencing, and the CAMCOR facility at the University of Oregon for the phage micrographs.
References
- [1].Genersch E. Honey bee pathology: current threats to honeybees and beekeeping. App Microbiol Biotechnol 2010; 87:87-97; PMID:20401479; http://dx.doi.org/19909971 10.1007/s00253-010-2573-8 [DOI] [PubMed] [Google Scholar]
- [2].De Graaf DC, Alippi AM, Antunez K, Aronstein KA, Budge G, De Koker D, De Smet L, Dingman DW, Evans JD, Foster LJ, Fünfhaus A, Garcia-Gonzalez E, Gregore A, Human H, Murray KD, Nguyen BK, Poppinga L, Spivak M, Engelsdorp D, Wilkins S, and Genersch E. Standard methods for American foulbrood research. J Apicult Res. 2012; 52(1):1-26; http://dx.doi.org/ 10.3896/IBRA.1.52.1.11 [DOI] [Google Scholar]
- [3].Genersch E. American Foulbrood in honeybees and its causative agent, Paenibacillus larvae. J Invertebr Pathol 2009; 103:S10-19; PMID:19909971; http://dx.doi.org/ 10.1016/j.jip.2009.06.015 [DOI] [PubMed] [Google Scholar]
- [4].Miyagi T, Peng CY, Chuang RY, Mussen EC, Spivak MS, Doi RH. Verification of oxytetracycline-resistant American foulbrood pathogen Paenibacillus larvae in the United States. J Invertebr Pathol 2000; 75:95-6; PMID:10631065; http://dx.doi.org/ 10.1006/jipa.1999.4888 [DOI] [PubMed] [Google Scholar]
- [5].Tian B, Fadhil NH, Powell JE, Kwong WK, Moran NA. Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honeybees. mBio 2012; 3(6):e00377-12; PMID:23111871; http://dx.doi.org/ 10.1128/mBio.00377-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Matsuzaki S, Uchiyama J, Takemura-Uchiyama I, Daibata M. Perspective: The age of the phage. Nature 2014; 509(7498):S9; PMID:24784429; http://dx.doi.org/ 10.1038/509S9a [DOI] [PubMed] [Google Scholar]
- [7].Hagens S, Loessner MJ. Application of bacteriophages for detection and control of foodborne pathogens. Appl Microbiol Biotechnol 2007; 76(3):513-519; PMID:17554535; http://dx.doi.org/ 10.1007/s00253-007-1031-8 [DOI] [PubMed] [Google Scholar]
- [8].Loc-Carrillo C, Abedon ST. Pros and cons of phage therapy. Bacteriophage 2011; 1(2):111-4; PMID:22334867; http://dx.doi.org/ 10.4161/bact.1.2.14590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chan BK, Abedon ST, Loc-Carrillo C. Phage cocktails and the future of phage therapy. Fut Microbiol 2013; 8(6):769-783; PMID:23701332; http://dx.doi.org/ 10.2217/fmb.13.47 [DOI] [PubMed] [Google Scholar]
- [10].Gochnauer TA. The isolation of a bacteriophage (bacterial virus) from Bacillus larvae. Bee World 1955; 36:101-3; http://dx.doi.org/ 10.1080/0005772X.1955.11094880 [DOI] [Google Scholar]
- [11].Gochnauer TA. Some properties of a bacteriophage from Bacillus larvae. J Invertebr Pathol 1970; 15:149-56; PMID:5435792; http://dx.doi.org/ 10.1016/0022-2011(70)90228-4 [DOI] [PubMed] [Google Scholar]
- [12].Valerianov T, Popova A, Toshkov AS. Isolation from the soil of a bacteriophage lysing Bacillus larvae. Acta Microbiol Virol Immunol 1976; 4:81-5; PMID:1007965 [PubMed] [Google Scholar]
- [13].Drobnikova V, Ludvik J. Bacteriophage of Bacillus larvae. J Apic Res 1982; 21:53-6; http://dx.doi.org/ 10.1080/00218839.1982.11100516 [DOI] [Google Scholar]
- [14].Benada O, Ludvik J, Drobnikova V. Morphology of a new bacteriophage isolated from Bacillus larvae. Folia Microbiol 1984; 29:520-1; http://dx.doi.org/ 10.1007/BF02873162 [DOI] [Google Scholar]
- [15].Dingman DW, Bakhiet N, Field CC, Stahly DP. Isolation of two bacteriophages from Bacillus larvae, PBL1 and PBL0.5, and partial characterization of PBL1. J Gen Virol 1984; 65:1101-5; PMID:6726188; http://dx.doi.org/ 10.1099/0022-1317-65-6-1101 [DOI] [PubMed] [Google Scholar]
- [16].Bakhiet N, Stahly DP. Properties of clear plaque mutants of the Bacillus larvae bacteriophages PBL0.5 and PBL 2. J Invertebr Pathol 1988; 52:78-83; http://dx.doi.org/ 10.1016/0022-2011(88)90105-X [DOI] [Google Scholar]
- [17].Campana CF, Bakhiet N, Stahly DP. Morphology of Bacillus larvae bacteriophage PBL3 and physical map of its DNA. J Invertebr Pathol 1991; 57:141-3; http://dx.doi.org/ 10.1016/0022-2011(91)90055-U [DOI] [Google Scholar]
- [18].Stahly DP, Alippi AM, Bakhiet N, Campana CF, Novak CC, Cox R. PPL1c, a virulent mutant bacteriophage useful for identification of Paenibacillus larvae subspecies larvae. J Invertebr Pathol 1999; 74:295-6; PMID:10534418; http://dx.doi.org/ 10.1006/jipa.1999.4893 [DOI] [PubMed] [Google Scholar]
- [19].LeBlanc L, Nezami S, Yost D, Tsourkas P, Amy PS. Isolation and characterization of a novel phage lysin active against Paenibacillus larvae, a honeybee pathogen. Bacteriophage 2015; 5(4):e1080787; PMID:26904379; http://dx.doi.org/ 10.1080/21597081.2015.1080787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yost DG, Tsourkas P, Amy PS. Experimental bacteriophage treatment of honeybees (Apis mellifera) infected with Paenibacillus larvae, the causative agent of American Foulbrood disease. Bacteriophage 2016; 6(1):e1122698; http://dx.doi.org/ 10.1080/21597081.2015.1122698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ghorbani-Nezami S, LeBlanc L, Yost DG, Amy PS. Phage therapy is effective in protecting honeybee larvae from American Foulbrood disease. J Insect Sci 2015; 15(1):84-9; PMID:26136497; http://dx.doi.org/ 10.1093/jisesa/iev051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Oliveira A, Leite M, Kluskens LD, Santos SB, Melo LDR, Azeredo J. The first Paenibacillus larvae bacteriophage endolysin (PlyPl123) with high potential to control American Foulbrood. PLoS One 2015; 11(2):e0150157; http://dx.doi.org/ 10.1371/journal.pone.0150157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Beims H, Wittman J, Bunk B, Sproer C, Rohde C, Gunther G, Rohde M, von der Ohe W, Steinert M. Paenibacillus larvae-directed bacteriophage HB10c2 and its application in American Foulbrood-affected honey bee larvae. Appl Environ Microbiol 2015; 81(16):5411-9; PMID:26048941; http://dx.doi.org/ 10.1128/AEM.00804-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Oliveira A, Melo LDR, Kropinski AM, Azeredo J. Complete genome sequence of the broad-host-range Paenibacillus larvae phage philBB_P123. Genome Announc 2013; 1(5):e00438-13; PMID:24009112; http://dx.doi.org/ 10.1128/genomeA.00438-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Carson S, Bruff E, DeFoor W, Dums J, Groth A, Hatfield T, Iyer A, Joshi K, McAdams S, Miles D, et al.. Genome sequences of six Paenibacillus larvae Siphoviridae phages. Genome Announc 2015; 3(3):e00101-15; http://dx.doi.org/ 10.1128/genomeA.00101-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tsourkas P, Yost D, Krohn A, Leblanc L, Zhang A, Stamereilers C, Amy PS. Complete genome sequences of nine phages capable of infecting Paenibacillus larvae, the causative agent of American Foulbrood disease of honeybees. Genome Announc 2015; 3(5):e01120-15; PMID:26472825; http://dx.doi.org/ 10.1128/genomeA.01120-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].White GF. The bacteria of the apiary with special reference to bee disease In: Bureau of Entomology, Technical Series. Washington, DC: US Department of Agriculture, 1906; 14:1-50. [Google Scholar]
- [28].Nelson DC, Schmelcher M, Rodriguez-Rubio L, Klumpp J, Pritchard DG, Dong S, Donovan DM. Endolysins as antimicrobials. Adv Virus Res 2012; 83:299-365; http://dx.doi.org/ 10.1016/B978-0-12-394438-2.00007-4 [DOI] [PubMed] [Google Scholar]
- [29].Payne KM, Hatfull GF. Mycobacteriophage endolysins: diverse and modular enzymes with multiple catalytic activities. PLoS One 2012; 7(3):e34052; PMID:22470512; http://dx.doi.org/ 10.1371/journal.pone.0034052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Walmagh M, Boczkowska B, Grymonprez B, Briers Y, Drulis-Kawa Z, Lavigne R. Characterization of five novel endolysins from Gram-negative infecting bacteriophages. Appl Microbiol Biotechnol 2013; 97(10):4369-75; PMID:22832988; http://dx.doi.org/ 10.1007/s00253-012-4294-7 [DOI] [PubMed] [Google Scholar]
- [31].Cotter PD, Ross RP, Hill C. Bacteriocins - a viable alternative to antibiotics? Nat Rev Microbiol 2013 Feb; 11(2):95-105; PMID:23268227; http://dx.doi.org/ 10.1038/nrmicro2937 [DOI] [PubMed] [Google Scholar]
- [32].Aunpad R, Panbangred W. Evidence for two putative holin-like peptides encoding genes of Bacillus pumilus strain WAPB4. Curr Microbiol 2012; 64(4):343-8; PMID:22231453; http://dx.doi.org/ 10.1007/s00284-011-0074-3 [DOI] [PubMed] [Google Scholar]
- [33].Anthony T, Chellappa GS, Rajesh T, Gunasekaran P. Functional analysis of a putative holin-like peptide-coding gene in the genome of Bacillus licheniformis AnBa9. Arch Microbiol 2010; 192(1):51-6; PMID:19967339; http://dx.doi.org/ 10.1007/s00203-009-0530-7 [DOI] [PubMed] [Google Scholar]
- [34].Hatfull GF, Jacobs-Sera D, Lawrence JG, Pope WH, Russell DA, Ko CC, Weber RJ, Patel MC, Germane KL, Edgar RH, et al.. Comparative genomic analysis of 60 Mycobacteriophage genomes: genome clustering, gene acquisition, gene size. J Mol Biol 2010; 397:119-43; PMID:20064525; http://dx.doi.org/ 10.1016/j.jmb.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pope WH, Bowman CA, Russell DA, Jacobs-Sera D, Asai DJ, Cresawn SG, Jacobs WR, Hendrix RW, Lawrence JG, Hatfull GF. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. Elife 2015; 4:e06416; PMID:25919952; http://dx.doi.org/ 10.7554/eLife.06416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Casjens Sr. Comparative genomics and evolution of the tailed-bacteriophages. Curr Opin Microbiol 2005; 8:451-8; PMID:16019256; http://dx.doi.org/ 10.1016/j.mib.2005.06.014 [DOI] [PubMed] [Google Scholar]
- [37].Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev 1992; 56(3):430-81; PMID:1406491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al.. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012; 28(12):1647-9; PMID:22543367; http://dx.doi.org/ 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Delcher AL, Harmon D, Kasif S, White O, Salzberg S. Improved microbial gene identification with GLIMMER. Nucl Acids Res 1999; 27(23):4636-41; PMID:10556321; http://dx.doi.org/ 10.1093/nar/27.23.4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Besemer J, Borodovsky M. GeneMark: Web software for gene finding in prokaryotes eukaryotes, and viruses. Nucl Acids Res 2005; 33(suppl 2):W451-4; http://dx.doi.org/ 10.1093/nar/gki487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lukashin AV, Borodovsky M. GeneMark.hmm: New solutions for gene finding. Nucl Acids Res 1998; 26(4):1107-15; PMID:9461475; http://dx.doi.org/ 10.1093/nar/26.4.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Krumsiek J, Arnold R, Rattei R. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 2007; 23(8):1026-28; PMID:17309896; http://dx.doi.org/ 10.1093/bioinformatics/btm039 [DOI] [PubMed] [Google Scholar]
- [43].Thompson JD, Higgins DG, Gibson TJ. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 1994; 22(22):4673-80; PMID:7984417; http://dx.doi.org/ 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al.. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011; 7(1):1-6; PMID:21988835; http://dx.doi.org/ 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JF, Geer LY, Geer RC, Gonzales NR, Gwadz M, et al.. CDD: specific functional annotation with the Conserved Domain Database. Nucl Acids Res 2009; 37(suppl 1):D205-10; PMID:18984618; http://dx.doi.org/ 10.1093/nar/gkn845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cresawn SG, Bogel M, Day N, Jacobs-Sera D, Hendrix RW, Hatfull GR. Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics 2011; 12:395-409; PMID:21219653; http://dx.doi.org/ 10.1186/1471-2105-12-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Merrill BD, Grose JH, Breakwell DP, Burnett SH. Characterization of Paenibacillus larvae bacteriophages and their genomic relationships to firmicute bacteriophages. BMC Genomics 2014; 15:745; PMID:25174730; http://dx.doi.org/ 10.1186/1471-2164-15-745 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



