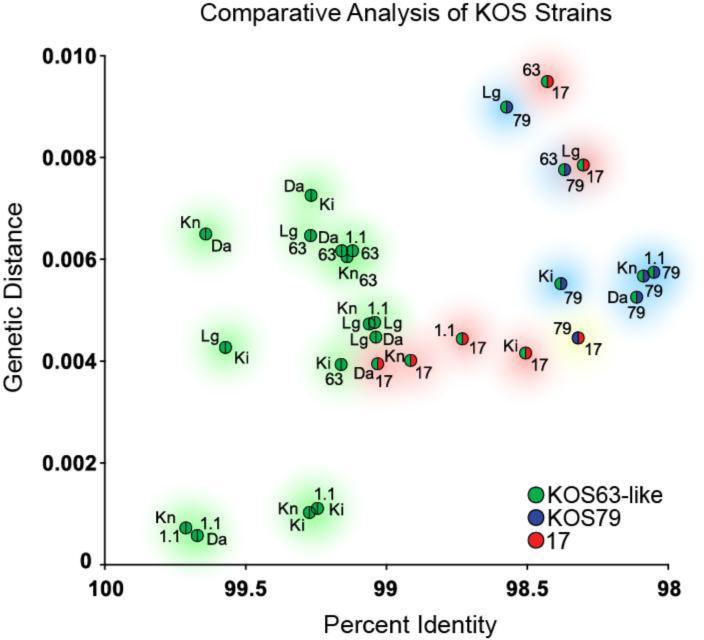
Figure 1.
Genome sequence comparisons reveal that KOS63 is as different from KOS79 as from the HSV-1 reference strain 17. The genome sequences of multiple variants of KOS63 (green) were compared to those of KOS79 (blue) and the HSV-1 reference strain 17 (red). For every possible pairwise combination of genomes, we calculated the genetic distance and the percent DNA identity (see Methods for details; see also Table 3). Points are color-coded to reflect the pairs in each comparison, with strain labels next to each point (see below for abbreviations). The x-axis reflects decreasing percent identity, while the y-axis reflects increasing genetic distance. In both cases, more disparate pairwise comparisons will plot further from the origin. Shaded circles provide a visual guide to the groups of pairwise comparisons. Green shading indicates pairwise combinations of KOS63-like strains, blue shading indicates combinations involving strain KOS79, and red shading indicates combinations involving strain 17. Yellow shading indicates the sole pairwise combination of strains 17 and KOS79. Trimmed genome sequences, lacking the terminal copies of the large repeats, were used for all comparisons to avoid giving undue weight to the repeats. Reiteration lengths in several published strains (32, 34) are artificially matched to strain 17, so these regions were removed from all comparisons as well (see Methods for details). Abbreviations are as follows: Da, KOSDavido from Davido and colleagues (32); Lg, KOSLarge from Szpara and colleagues (29); Ki, KOSKinchington from Kinchington and colleagues (33); Kn, KOSKnipe and 1.1, KOS1.1, from Knipe and colleagues (34); 63, KOS63 as described by Dix and colleagues (22), with genome sequenced here; 79, KOS79 as described by Dix and colleagues (22), with genome sequenced here; 17, HSV-1 reference strain 17, as initially published by McGeoch and colleagues (20, 21), and revised by Davison and colleagues (76). All GenBank accessions are listed in Table 1.