Fig. 1.
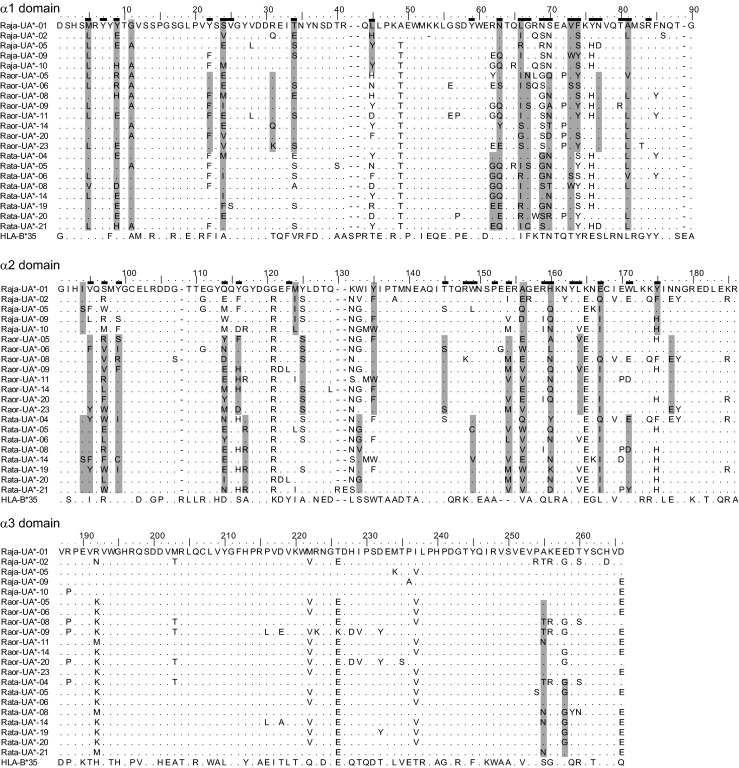
Amino acid alignment of selected MHC class I variants from R. japonica (Raja), R. ornativentris (Raor) and R. t. tagoi (Rata). Alignment is divided into the three domains studied. Codon sites shaded in grey are detected as under positive selection independently in each species by at least two of four methods used (omegaMap, FEL, REL and MEME); referred to as positively selected sites (PSS). Putative PBR sites inferred from humans (Bjorkman et al. 1987; Saper et al. 1991) are indicated by black bars above the leading sequence. Amino acid sequence for a well-characterised HLA allele, HLA-B*35, is also included (HLA-B*35:01:01:01, IMGT/HLA acc. no. HLA00237)
