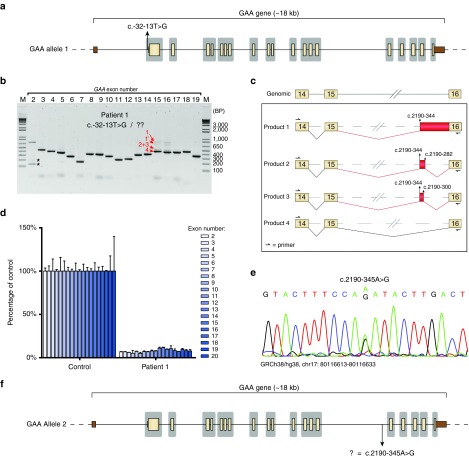
Figure 1.
Identification of a deep intronic pathogenic variant in patient 1 using the splicing assay. (a) Scaled cartoon of acid α-glucosidase (GAA) allele 1 from patient 1 carrying the c.-32-13T>G (IVS1) variant. Coding exons are indicated in yellow, untranslated regions in brown. Lines represent introns. Areas in gray indicate the regions that are sequenced as part of standard diagnostic practice. (b) Flanking exon reverse-transcriptase-polymerase chain reaction (RT-PCR) of all GAA coding exons. Asterisks indicate known aberrant splicing products caused by the IVS1 variant. Products 1–4 indicate novel mRNA products. Product 1′ refers to a secondary structural variant of product 1 (see Supplementary Figure S1b). (c) Cartoon of splicing products 1–4. Boxes indicate exonic regions, (dashed) lines represent intronic regions. Aberrant splicing events are indicated in red. (d) Exon internal RT-quantitative PCR analysis of GAA exons 2–20. Data are normalized for β-actin and for a healthy control. Data represent means of three technical replicates ± SD. (e) Sequence analysis of genomic DNA of the region surrounding the c.2190–344 cryptic splice acceptor site. (f) Cartoon of GAA allele 2 showing the location of the c.2190-345A>G variant.