Abstract
Aim:
Fascioliasis is one of the most zoonotic diseases with global extension. As the epidemiological distribution of Fasciola may lead to various genetic patterns of the parasite, the aim of this study is to identify Fasciola hepatica based on spermatogenesis, and phylogenetic analysis using mitochondrial (nicotiamide adenine dinucleotide dehydrogenase subunit I [ND1] and cytochrome oxidase subunit I) gene marker.
Materials and Methods:
In this study, 90 F. hepatica collected from 30 cattle at slaughterhouse located in three different geographical locations in the North-East of Iran were evaluated based on spermatogenetic ability and internal transcribed spacer 1 gene restriction fragment length polymorphism pattern. Genetic diversity and phylogenetic relationship using mtDNA gene marker for the isolates from the North-East of Iran, and other countries were then analyzed.
Results:
Partial sequences of mtDNA showed eight haplotypes in both genes. The phylogenic analysis using neighbor joining as well as maximum likelihood methods showed similar topologies of trees. Pairwise fixation index between different F. hepatica populations calculated from the nucleotide data set of ND1 gene are statistically significant and show the genetic difference.
Conclusion:
F. hepatica found in this region of Iran has different genetic structures through the other Fasciola populations in the world.
Keywords: Cytochrome oxidase subunit I, Fasciola hepatica, Iran, molecular characterization, nicotiamide adenine dinucleotide dehydrogenase subunit I, phylogenic
Introduction
Fasciolosis is a zoonotic parasitic disease that occurring in domestic grazing animals, affected on livestock economy by decrease in output products. The financial burden is projected to be around two million US dollars annually [1]. The causative species most commonly implicated of fasciolosis are Fasciola hepatica and Fasciola gigantica that F. hepatica is distributed worldwide, although F. gigantica is fixed to warm parts and has been present in Africa along with South and Southeast Asia [2,3]. The two species have now been notable on the morphological criteria such as body size and shape; nevertheless, these requirements are not generally trusted due to the morphological variety within the species [4].
On the other hand, these species are meiotically diploid and produce sperm and store in the seminal vesicle (spermic), but some Asian Fasciola forms are meiotically dysfunctional and cannot produce sperm (aspermic) [5,6].
Two species may be identified using molecular methods by nucleotide sequences of nuclear ribosomal central internal transcribed spacer 1 (ITS1) and ITS2 [7-9]. In addition, DNA sequences of mitochondrial nicotiamide adenine dinucleotide dehydrogenase subunit I (ND1) and cytochrome oxidase subunit I (CO1) genes have been applied to analyze intraspecific phylogenic relations of Fasciola spp. [9,10].
It’s been thought that F. hepatica started in Europe and circulation of flukes depended on migration of livestock with individual colonizers [10].
Iran is a thorough country with various geographical ecology and existence of both species from cattle observed. There are several studies from Iran ruminant fascioliasis, especially in cattle and buffaloes centered on geography and weather variability [11]. Although some molecular studies have been conducted in several areas for genotyping of Fasciola species without a study on spermatogenetic ability in Iran [11], there is no useful finding based on populace design and genetic modifications of F. hepatica Iran.
The objective of this study was not only to identify of F. hepatica based on spermatogenesis and ITS1 marker by PCR-restriction fragment length polymorphism (RFLP), but also analyze their phylogenetic relationship with population from different parts of the world in the North East of Iran using ND1 and CO1 as mitochondrial makers with available GenBank records. Furthermore, genetic variability of F. hepatica foci of this region of Iran inferred by mitochondrial DNA sequences to get the parasitic gene flow among different populations was evaluated.
Materials and Methods
Ethical approval
This research was approved by the Faculty of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
F. hepatica collections and spermatogenetic ability
A total of 90 adults of Fasciola specimens were collected from the bile ducts 30 cattle at slaughterhouse situated in three different geographical locations in the North East of Iran from January to September 2015. These cattle (Bus taurus) traditionally were nurtured. There are two provinces in the North East of Iran (North and Razavi Khorasan) bordering Turkmenistan and Afghanistan country (Figure-1). Fasciola flukes were washed in 0.9% saline solution and fixed in 70% ethanol between two glass slides and transported to the laboratory for further studies. The seminal vesicles of fixed specimens in the anterior part of body were removed and stained with hematoxylin–carmine solution, and then observed under a visual microscope to examine for the existence of sperm [12,13]. The posterior parts, excluding the uterus, which might contain sperm from other individuals, were used for total DNA extraction.
Figure 1.
The provinces of the North and Razavi Khorasan in Iran.
DNA extraction and amplification
Total DNA was extracted from each fluke with a High Pure PCR Template Preparation Kit (Dynabio®, Takapouzist, Iran), according to the manufacturer’s instructions and stored at −20°C until use. DNA fragments of each target region were amplified by polymerase chain reaction (PCR) using a pair primer shown in Table-1 [14]. The total volume of reaction was 15 μl containing 1.5 μl DNA template, 5 μl distilled water, 10 pmol of each primers, and 7.5 μl master mix (amplicon). Reaction cycles consisted of an initial denaturing step at 94°C for 90 s, followed by 35 cycles at 94°C for 90 s, 53°C (ITS1) or 55°C (ND1 and CO1) for 90 s, and 72°C for 120 s, with a final extension at 72°C for 10 min using a gradient thermocycler. DNA fragments were analyzed by 1.5% agarose gel electrophoresis [9].
Table-1.
The name and sequences of the primers used in this research.
| Gene | Name | Sequence |
|---|---|---|
| ITS1 | ITS1-F | 5′-TTGCGCTGATTACGTCCCTG-3′ |
| ITS1-R | 5′-TTGGCTGCGCTCTTCATCGAC-3′ | |
| ND1 | Ita 10 | 5′-AAGGATGTTGCTTTGTCGTGG-3′ |
| Ita 2 | 5′-GGAGTAC GGTTACATTCACA-3′ | |
| CO1 | Ita 8 | 5′-ACGTTGGATCATAAGCGTGT-3′ |
| Ita 9 | 5′-CCTCATCCAACATAACCTCT-3′ |
ITS1=Internal transcribed spacer 1, ND1=Nicotiamide adenine dinucleotide dehydrogenase subunit I, CO1=Cytochrome oxidase subunit I
RFLP of amplified DNA (PCR-RFLP)
The ampilcons of the ITS1 region were examined by the PCR-RFLLP method. Briefly, the reaction level of 10 μL contained 5 μL of PCR products with approximately 680-bp fragments, 1 U of the RsaI restriction enzyme, and 1 μL of manufacturer supplied reaction buffer (Cinagen®, Iran). After incubation at 37°C for 3 h and heat inactivation of RsaI at 65°C for 15 min, the digested DNA samples were analyzed by gel electrophoresis [8].
Sequences and phylogenetic analysis
Products of ND1 and CO1 of isolates sequenced by Bioneer Company utilizing the same primers, which were found in the PCR. The sequences were aligned and in contrast to those of existing sequences from the region, linked to Fasciola spp., available in the GenBank, utilizing the Chromas 2.2 and multiple alignments were performed with data linked to F. hepatica from Iran and other countries deposited in GenBank. Phylogenic analyses predicated on NDI and COI sequence data were conducted by maximum likelihood using MEGA6 [15]. All characters were run unordered and equally weighted. Alignment gaps were treated as missing data. Bootstrap analyses were conducted using 1000 replicates.
Genetic diversity indices
Diversity indices (Haplotype diversity; Hd and nucleotide diversity: π) and neutrality indices (Tajima’s D and Fu’s Fs tests) were estimated by DnaSP software version 5.10 [16]. The people genetic structure was analyzed by Arlequin version 3.11 [17]. The degree of gene flow (gene migration) on the list of populations was evaluated utilizing a pairwise fixation index (Fst) [18].
Results
All 90 sample specimens had many normal sperms in the seminal vesicles and were spermic species. Based on the RFLP fragment pattern in ITS1 region, they exhibited the F. hepatica type. ND1 fragments (approximately 535 bp) and CO1 fragments (approximately 438 bp) were amplified for several specimens. Partial sequences of ND1 and CO1 showed 26 and 11 variable sites, respectively, and also yielded eight haplotypes in both genes and high diversity indices in ND1 gene (Table-2). The nucleotide sequences for every haplotype were deposited in GenBank under following accession numbers: KX021280-KX021299. The profiles of F. hepatica haplotypes based on ND1 and CO1 genes and type of nucleotides in the North East of Iran shown in Table-3. Phylogenic analyses based on ND1 and CO1 sequence data were conducted by neighbor-joining (NJ) using MEGA6 with F. gigantica designated as an outgroup showed in Figures-2 and 3. Pairwise fixation index (Fst values) between different F. hepatica populations calculated from the nucleotide data group of ND1 gene are statistically significant and show the genetic difference in pairwise population (Table-4).
Table-2.
Haplotype diversity and nucleotide diversity of Fasciola fluke in North East of Iran based on ND1 gene.
| Species | Population | Diversity indices | Neutrality indices | ||||
|---|---|---|---|---|---|---|---|
| n | Hn | Hd±SD | π | Tajima’s D | Fu’s Fs statistic | ||
| F. hepatica | North East of Iran | 90 | 14 | 0.997±0.003 | 0.01552 | −1.3724* | −2.59 |
Statistical significance: Not significant, p>0.1. Hn=Number of haplotypes, Hd=Haplotype diversity, Nd=Nucleotide diversity, SD=Standard deviation, F. hepatica=Fasciola hepatica, ND1=Nicotiamide adenine dinucleotide dehydrogenase subunit I
Table-3.
Profiles of F. hepatica haplotypes and their accession no in the North East of Iran.
| Location (Province) | Bojnurd and Shirvan (North Khorasn) | Maneh and Samalghan (North Khorasn) | Ghochan (Razavi Khorasn) |
|---|---|---|---|
| Host (number) | 12 | 8 | 10 |
| Number of flukes | 38 | 25 | 27 |
| Sperm in seminal vesicles | + | + | + |
| ITS1-RFLP | F. hepatica | F. hepatica | F. hepatica |
| Haplotype (accession no.) | |||
| ND1 | B1ND (KX021280) B2ND (KX021281) B3ND (KX021282) B4ND (KX021283) B5ND (KX021284) |
B3ND (KX021282) B6ND (KX021285) |
B6ND (KX021285) B7ND (KX021286) B8ND (KX021287) |
| CO1 | B1CX (KX021290) B2CX (KX021291) B3CX (KX021292) B4CX (KX021293) |
B5CX (KX021294) | B6CX (KX021295) B7CX (KX021296) B8CX (KX021297) |
ITS1=Internal transcribed spacer 1, ND1=Nicotiamide adenine dinucleotide dehydrogenase subunit I, CO1=Cytochrome oxidase subunit I, RFLP=Restriction fragment length polymorphism, F. hepatica=Fasciola hepatica
Figure 2.
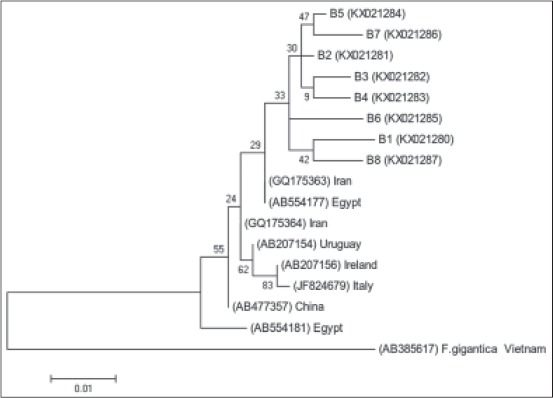
Phylogenetic relationship based on nicotiamide adenine dinucleotide dehydrogenase subunit I sequences of Fasciola hepatica from the North East of Iran. The tree constructed by MEGA6 using neighbor-joining analysis. Scale bars indicated nucleotide substitutions per site. Fasciola gigantica was used as outgroup.
Figure 3.
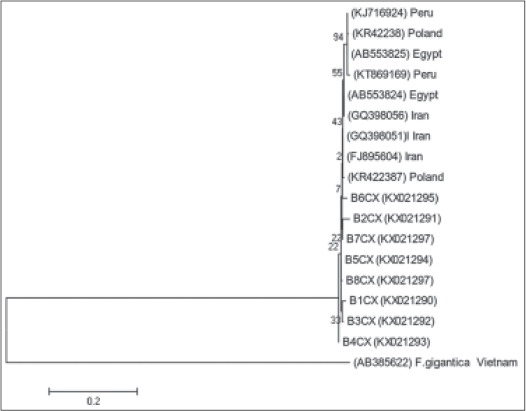
Phylogenetic relationship based on cytochrome oxidase subunit I sequences of Fasciola hepatica from the North East of Iran. The tree constructed by MEGA6 using neighbor-joining analysis. Scale bars indicated nucleotide substitutions per site. Fasciola gigantica was used as outgroup.
Table-4.
Pairwise fixation index (Fst values) between different F. hepatica populations calculated from the nucleotide data set of ND1 gene.
| Population | Population | ||||
|---|---|---|---|---|---|
| Iran (this study) | Egypt | Peru | Asia* | Europe** | |
| Iran (this study) | - | ||||
| Egypt | 0.98329 | - | |||
| Peru | 0.98516 | 0.98892 | - | ||
| Asia* | 0.89128 | 0.89282 | 0.01768 | - | |
| Europe** | 0.78107 | 0.78042 | 0.00250 | −0.06162 | - |
Asia: China, Thailand, Japan,
Europe: Italy, Poland. All values are statistically non-significant (p>0.05). ND1=Nicotiamide adenine dinucleotide dehydrogenase subunit I, F. hepatica=Fasciola hepatica
Discussion
Microscopical observation after staining revealed that spermic Fasciola occurred in the North East of Iran. There are always a few studies in Iran that concentrate on spermatogenesis in Fasciola sp. Ashrafi et al. reported F. hepatica, F. gigantica and intermediate forms in the endemic region of the North East of Iran (Gilan province) based on morphological and phenotypic analysis of Fasciola flukes [4]. In this study, all of the samples were spermic F. hepatica with spermatogenetic ability.
Epidemiological patterns are crucial and the important factors in differentiation of Fasciola species. Despite the fact that phenotypic criteria have been considered to be one of the useful criteria for discrimination of species in Fasciola [19], but molecular approaches clarify of global genetic diversity and distinguish intraspecific relations. RFLP methods using ITS regions were used to identify the Fasciola species and has been extensively validated [20,21]. This method has been used in a number of studies in Iran [22,23], but studies showed that molecular phylogeny with mtDNA, including ND1 and CO1, could be effectively useful for proper differentiation of haplotypes [9,14].
The Fst values showed that F. hepatica population in three continents was genetically different from one another centered on NDI region (Table-3). These results could be related to the current presence of different haplotypes of investigated populations. Furthermore, this implies that here’s no transfer of alleles in one population to another population through immigration of F. hepatica.
The phylogenic analysis using NJ in addition to ML methods showed similar topologies of trees. Some reports from Iran were detected Fasciola sp., and constructed phylogenic trees using nuclear rDNA and declare that because of variation in this region; it’s inadequate to separate your lives of Fasciola species for resolving the taxonomic problem [24-26].
ND1 haplotypes of this study show a high range of diversity, but they belonged to one clade in shown phylogenetic tree. The phylogenetic trees showed relationship of isolated of other regions in Iran, but more studies and the population analysis need to get exact classification pattern.
Conclusion
This study demonstrated that F. hepatica found in this region of Iran has different genetic structures through the other Fasciola populations in accordance with pairwise fixation index, but to complete and find genetic diversity other molecular studies from another region of Iran is necessary.
Authors’ Contributions
SaberR and SR together have designed, planned and conducted this research. AH, MFH, and AS assisted in the execution. KA collected specimens. All authors have read and approved the final version of the manuscript.
Acknowledgments
This article has been extracted from the Ph.D. thesis written by Saber Raeghi in Department of Parasitology, Faculty of Medicine at Shahid Beheshti University of Medical Sciences (Registration No.: 359).
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Mas-Coma S, Bargues M. D, Valero M. A. Fascioliasis and other plant-borne trematode zoonoses. Int. J. Parasitol. 2005;35(11):1255–1278. doi: 10.1016/j.ijpara.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 2.Olaechea F, Lovera V, Larroza M, Raffo F, Cabrera R. Resistance of Fasciola hepatica against triclabendazole in cattle in Patagonia (Argentina) Vet. Parasitol. 2011;178(3):364–366. doi: 10.1016/j.vetpar.2010.12.047. [DOI] [PubMed] [Google Scholar]
- 3.Rokni M. B, Lotfy W. M, Ashrafi K, Murrell K. D. Fasciolosis in the MENA region. In: Neglected Tropical Diseases-Middle East and North Africa. Springer, Vienna. 2014:59–90. [Google Scholar]
- 4.Ashrafi K, Valero M. A, Panova M, Periago M. V, Massoud J, Mas-Coma S. Phenotypic analysis of adults of Fasciola hepatica, Fasciola gigantica and intermediate forms from the endemic region of Gilan, Iran. Parasitol. Int. 2006;55(4):249–260. doi: 10.1016/j.parint.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Peng M, Ichinomiya M, Ohtori M, Ichikawa M, Shibahara T, Itagaki T. Molecular characterization of Fasciola hepatica Fasciola gigantica and aspermic Fasciola sp. in China based on nuclear, and mitochondrial DNA. Parasitol. Res. 2009;105(3):809–815. doi: 10.1007/s00436-009-1459-0. [DOI] [PubMed] [Google Scholar]
- 6.Terasaki K, Itagaki T, Shibahara T, Noda Y, Moriyama-Gonda N. Comparative study of the reproductive organs of Fasciola groups by optical microscope. J. Vet. Med. Sci. 2001;63(7):735–742. doi: 10.1292/jvms.63.735. [DOI] [PubMed] [Google Scholar]
- 7.Adlard R. D, Barker S. C, Blair D, Cribb T. H. Comparison of the second internal transcribed spacer (ribosomal DNA) from populations and species of Fasciolidae (Digenea) Int. J. Parasitol. 1993;23(3):423–425. doi: 10.1016/0020-7519(93)90022-q. [DOI] [PubMed] [Google Scholar]
- 8.Ichikawa M, Itagaki T. Discrimination of the ITS1 types of Fasciola spp. based on a PCR-RFLP method. Parasitol. Res. 2010;106(3):757–761. doi: 10.1007/s00436-010-1724-2. [DOI] [PubMed] [Google Scholar]
- 9.Itagaki T, Kikawa M, Sakaguchi K, Shimo J, Terasaki K, Shibahara T, Fukuda K. Genetic characterization of parthenogenic Fasciola sp in Japan on the basis of the sequences of ribosomal and mitochondrial DNA. Parasitology. 2005:679–685. doi: 10.1017/S0031182005008292. [DOI] [PubMed] [Google Scholar]
- 10.Mas-Coma S, Valero M. A, Bargues M. D. Fasciola, lymnaeids and human fascioliasis, with a global overview on disease transmission, epidemiology, evolutionary genetics, molecular epidemiology and control. Adv. Parasitol. 2009;69:41–146. doi: 10.1016/S0065-308X(09)69002-3. [DOI] [PubMed] [Google Scholar]
- 11.Ashrafi K. The status of human and animal Fascioliasis in Iran: A narrative review article. Iran. J. Parasitol. 2015;10(3):306. [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi K, Ichikawa-Seki M, Mohanta U. K, Singh T. S, Shoriki T, Sugiyama H, Itagaki T. Molecular phylogenetic analysis of Fasciola flukes from Eastern India. Parasitol. Int. 2015;64(5):334–338. doi: 10.1016/j.parint.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Mohanta U. K, Ichikawa-Seki M, Shoriki T, Katakura K, Itagaki T. Characteristics and molecular phylogeny of Fasciola flukes from Bangladesh, determined based on spermatogenesis and nuclear and mitochondrial DNA analyses. Parasitol. Res. 2014;113(7):2493–2501. doi: 10.1007/s00436-014-3898-5. [DOI] [PubMed] [Google Scholar]
- 14.Itagaki T, Kikawa M, Terasaki K, Shibahara T, Fukuda K. Molecular characterization of parthenogenic Fasciola sp in Korea on the basis of DNA sequences of ribosomal ITS1 and mitochondrial NDI gene. J. Vet. Med. Sci. 2005;67(11):1115–1118. doi: 10.1292/jvms.67.1115. [DOI] [PubMed] [Google Scholar]
- 15.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rozas J, Sánchez-DelBarrio J. C, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19(18):2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 17.Excoffier L, Lischer H. E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under linux and windows. Mol. Ecol. Resour. 2010;10(3):564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 18.Reynolds J, Weir B. S, Cockerham C. C. Estimation of the coancestry coefficient: Basis for a short-term genetic distance. Genetics. 1983;105(3):767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Periago M. V, Valero M. A, Panova M, Mas-Coma S. Phenotypic comparison of allopatric populations of Fasciola hepatica and Fasciola gigantica from European and African bovines using a computer image analysis system (CIAS) Parasitol. Res. 2006;99(4):368–378. doi: 10.1007/s00436-006-0174-3. [DOI] [PubMed] [Google Scholar]
- 20.Chaichanasak P, Ichikawa M, Sobhon P, Itagaki T. Identification of Fasciola flukes in Thailand based on their spermatogenesis and nuclear ribosomal DNA, and their intraspecific relationships based on mitochondrial DNA. Parasitol. Int. 2012;61(4):545–549. doi: 10.1016/j.parint.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto K, Watanobe T, Liu C. X, Init I, Blair D, Ohnishi S, Agatsuma T. Mitochondrial DNA and nuclear DNA indicate that the Japanese Fasciola species is F. gigantica. Parasitol. Res. 1997;83(3):220–225. doi: 10.1007/s004360050237. [DOI] [PubMed] [Google Scholar]
- 22.Rokni M. B, Mirhendi H, Mizani A, Mohebali M, Sharbatkhori M, Kia E. B, Abdoli H, Izadi S. Identification and differentiation of Fasciola hepatica and Fasciola gigantica using a simple PCR-restriction enzyme method. Exp. Parasitol. 2010;124(2):209–213. doi: 10.1016/j.exppara.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 23.Shafiei R, Sarkari B, Moshfe A. A consistent PCR-RFLP assay based on ITS-2 ribosomal DNA for differentiation of Fasciola species. Iran. J. Basic Med. Sci. 2013;16(12):1266–1269. [PMC free article] [PubMed] [Google Scholar]
- 24.Aryaeipour M, Rouhani S, Bandehpour M, Mirahmadi H, Kazemi B, Rokni M. B. Genotyping and phylogenetic analysis of Fasciola spp. Isolated from sheep and cattle using PCR-RFLP in Ardabil province, Northwestern Iran. Iran. J. Public Health. 2014;43(10):1364–1371. [PMC free article] [PubMed] [Google Scholar]
- 25.Galavani H, Gholizadeh S, Tappeh K. H. Genetic characterization of Fasciola isolates from West Azerbaijan province Iran based on ITS1 and ITS2 sequence of ribosomal DNA. Iran. J. Parasitol. 2016;11(1):52–64. [PMC free article] [PubMed] [Google Scholar]
- 26.Yakhchali M, Malekzadeh-Viayeh R, Imani-Baran A, Mardani K. Morphological and molecular discrimination of Fasciola species isolated from domestic ruminants of Urmia City, Iran. Iran. J. Parasitol. 2015;10(1):46–55. [PMC free article] [PubMed] [Google Scholar]


