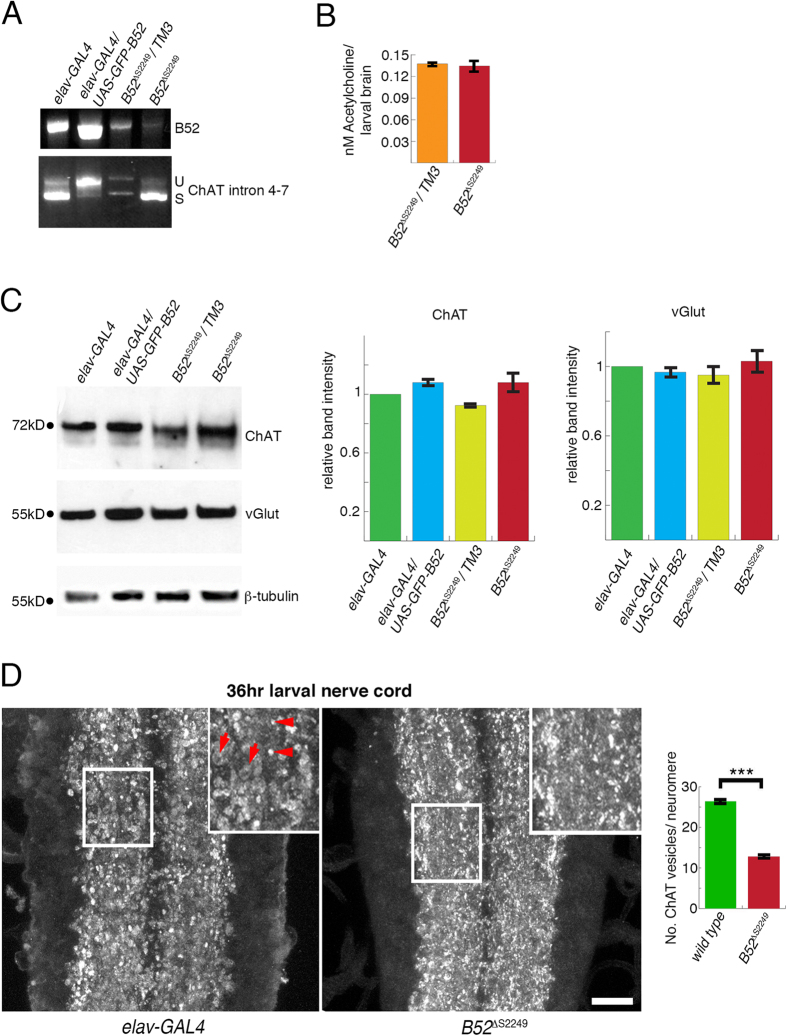
Figure 6. Gain but not loss of B52 affects splicing of ChAT.
Genotypes are indicated at the top or bottom of each panel: elav-GAL4 and B52ΔS2249/TM3, control; elav-GAL4/UAS-GFP-B52, gain of B52; UAS-BBS/+; elav-GAL4/+, B52 depletion; B52ΔS2249, B52 homozygous mutant. (A) Top panel: In 36 h larvae, increased neuronal expression of B52 (elav-GAL4/UAS-GFP-B52) raises the amount of B52 RNA and loss of B52 in heterozygous (B52Δ2249/TM3) and homozygous larvae (B52Δ2249) reduces B52 mRNA. In homozygous mutants B52 mRNA is barely detectable. Bottom panel: Gain of B52 mRNA results in a reduced efficiency of splicing of Intron 4–7 of ChAT. Whereas loss of B52 shows the opposite effect and splicing of intron 4–7 in B52Δ2249 mutant larvae seems as efficient as in controls (elav-GAL4). Images are cropped. For uncut images see Supplementary Fig. S4. (B) Loss of B52 does not change Acetylcholine synthesis in larval brains. Average acetycholine level per larval brain is shown (n = 2 experiments, 10 larval brains/experiment, bars are standard error). (C) Neither gain nor loss of B52 changes levels of ChAT protein (top) or vGlut levels (middle). Tubulin was used a loading control (bottom). Band intensity was adjusted to loading controls and normalized to elav-GAL4 control. Graphs represent band intensity for ChAT and vGlut as indicated on top of each graph. Three independent lysates were prepared, separated by SDS gelectrophoresis and blotted. Error bars show standard error. Images of blots are cropped. For uncut images see Supplementary Fig. S4. (D) Loss of B52 does not cause ectopic or increased expression of ChAT. ChAT in control larval CNS (left) and B52 homozygous mutants (middle) can be mainly detected along the longitudinal axon tracts. Yet, in control embryos, ChAT is present in structures resembling neuronal cell bodies (arrow, inset) as well as in small vesicles (arrowhead). In mutants, ChAT is limited to vesicles. Insets are 1.6x magnifications of framed area. Graph (right) represents the number of ChAT positive vesicles/neuromere. Vesicles in 2nd and 3rd abdominal neuromere with a diameter above 1 μm were counted. Wild type: 26.43 vesicles per segment, n = 18; B52ΔS2249: 12.78 vesicles/neuromere, n = 14. p > 0.001; Bars represent standard error. Horizontal views, Anterior is up. Scale: 10 μm (16 μm in inset).