Fig. 3.
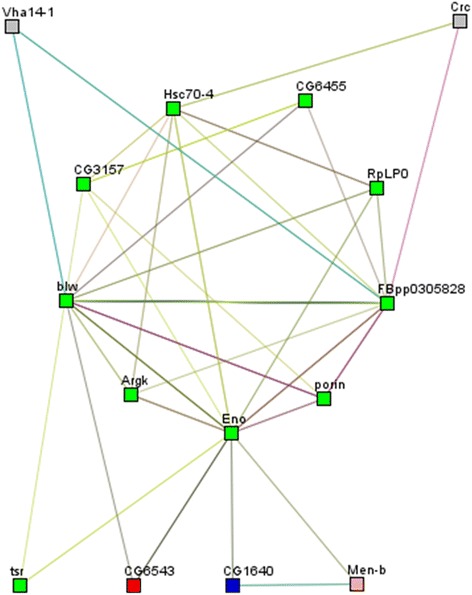
Differentially expressed proteins in the infected versus uninfected mosquito brain. Their interaction networks are depicted by STRING and visualized by Medusa. Each node represents the upregulated and downregulated proteins. The edges represent putative protein interactions recorded or predicted by STRING. The graph was obtained using the spectral clustering option, with five clusters. Node rectangles are coloured and the edges predicted functional links. Neighborhood (green lines), Gene Fusion (red lines), Co-occurrence (blue lines), Co-expression (grey lines), Experiments (purple lines), Databases (light blue lines), Textmining (light green lines) and Homology (light grey lines). Abbreviations: CG6543, enolyl-Coa hydratase; CG32683, HSP70; Men-b, malate dehydrogenase; Sap47, synapse-associated protein; CG14715, peptidyl-prolyl cis-trans isomerase; Eno, enolase; Porin, VDAC porin; Crc, calreticulin; Argk, arginine kinase; FBpp0305828, ATP synthase β; CG1640, alanine transaminase; CG6455, mitofilin; Vha14-1, V-type proton ATPase F; Tsr, cofilin; Hsc70-4, HSP70; CG3157, tubulin; GTPase, Gbeta13F protein G β-1; RpLP0, 60s Ribosomal LP0; Blw, F-type H+ transporting ATPase α
