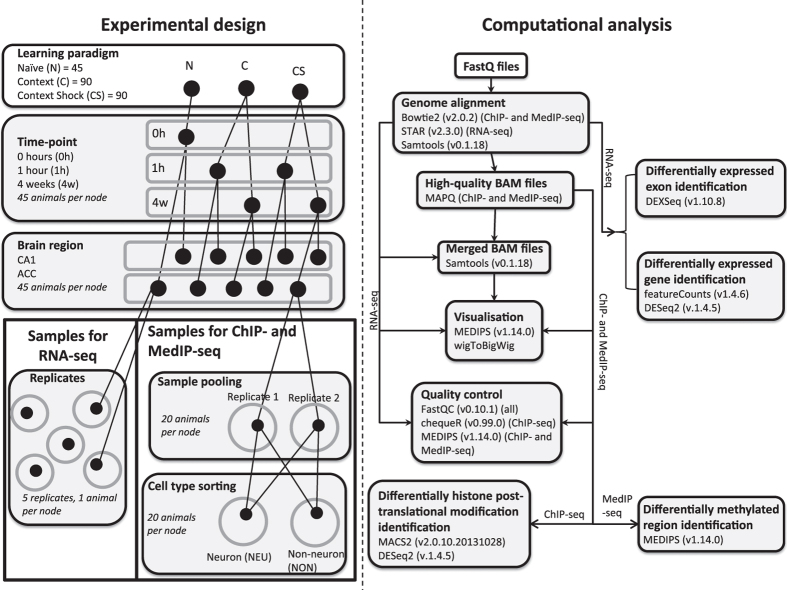
Figure 1. Overview of experimental design and computational analysis workflow.
Left; experimental design for the generation of samples to be sequenced with ChIP-, MeDIP- and RNA-seq. The mice are trained under different conditions (naïve-N, context-C or context shock-CS) and are sacrificed at different time-points after training (0 h for naïve, 1 h or 4 weeks for context and context-shock). The ACC and CA1 brain regions are then taken from each mouse. For samples that would be used for RNA-sequencing, each tissue sample is used to produce one replicate, resulting in 5 replicates for each combination of condition, time-point and tissue. For samples that would be sequenced with ChIP- and MeDIP-seq, 20 tissue samples are pooled for each tissue to generate two replicates, and the cells within the tissues are sorted by their type (neuronal-NEU or non-neuronal, -NON). Right; computational analysis used to process the ChIP-, MeDIP- and RNA-seq data. The main analysis steps involved alignment of the FastQ reads to the mouse genome, visualizing the data, establishing the quality of the data and running differential enrichment/expression analyses; for more information, see ‘Methods’ section.