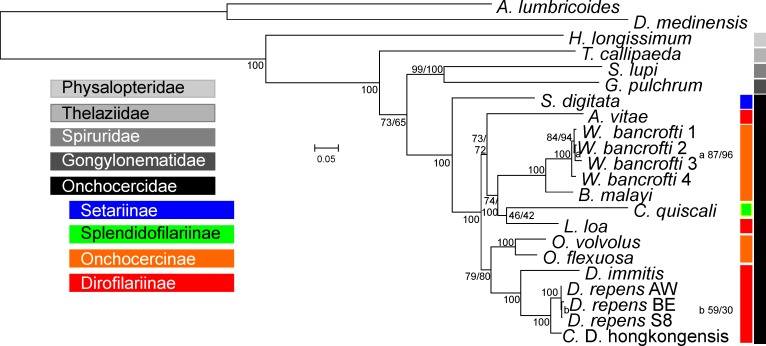
Fig 2. Phylogenetic analysis of spirurid nematodes using complete mitochondrial genomes.
The mitochondrial genomes of Dirofilaria repens (from top to bottom: KX265048, KX265047, KX265049), Dirofilaria hongkongensis (KX265047), Dirofilaria immitis, Onchocerca volvulus, Onchocerca flexuosa, Wuchereria bancrofti (from top to bottom: JN367461, JF775522, JQ316200, HQ184469), Brugia malayi, Chandlerella quiscali, Acanthocheilonema vitae, Setaria digitate, Heliconema longissimum, Spirocerca lupi, Thelazia callipedia and Gongylonema pulchrum were included in the analysis. The mitochondrial genomes of Ascaris lumbricoides and Dracunculus medinensis, clade III parasitic nematode not belonging to the Spirurida were used as outgroup. Concatenated alignments were analyzed with RAxML 8.1.11 with optimized models for each gene partition using the rapid bootstrapping algorithm. The Shimodaira-Hasegawa likelihood ratio test as implemented in RAxML was used as an alternative, unrelated method to obtain support values for all individual internal notes of this tree. Support values obtained by bootstrapping and by the likelihood ratio test are provided before and after the slash. If only one number is shown, the results of the tests were identical. Small letters were used if the space was too limited to write the support values directly at the branch. At the right of the scheme, different “classical” families within the Spirurida as defined by morphological criteria [81] are indicated as bars in different gray scales. As colored bars, different “classical” subfamilies within the Onchocercidae are indicated.