Abstract
Complement dependent cytotoxicity (CDC) is an important mechanism of action for monoclonal antibodies (mAb) used in the treatment of chronic lymphocytic leukemia (CLL). We hypothesized that alemtuzumab (ALM) mediated CDC would be increased by addition of ofatumumab (OFA). CLL cells from 21 previously untreated patients with progressive disease were tested in vitro for mAb binding, complement activation, and CDC. The subpopulation of CDC resistant CLL cells was examined for levels of C3b and C5b-9 binding, and expression of complement regulatory proteins. OFA significantly increased complement activation and CDC in ALM-treated CLL cells suggesting that combining ALM and OFA could improve clinical outcome in patients with CLL. Approximately 10% of CLL cells were resistant to CDC because of lower levels of complement activation or decreased cytotoxicity of activated complement. Improvement of clinical responses will require determining the mechanisms of CDC resistance and developing methods to overcome this problem.
Keywords: Chronic lymphocytic leukemia (CLL), complement dependent cytotoxicity, resistance, alemtuzumab, ofatumumab, rituximab
Introduction
Monoclonal antibodies (mAb) that target certain proteins on B cells are valuable drugs for the treatment of chronic lymphocytic leukemia (CLL). The CD52 specific mAb alemtuzumab (ALM) is highly effective in the treatment of CLL and is especially important in the management of patients with TP53 defective or purine analogue refractory disease [1]. Rituximab (RTX, specific for CD20) based chemoimmunotherapy has markedly increased response rates in the treatment of CLL [2–4] and addition of RTX to fludarabine and cyclophosphamide increases overall survival after initial treatment of progressive CLL [5]. The recently FDA-approved human anti-CD20 mAb ofatumumab (OFA) has appreciable activity in the treatment of CLL [6] and could be an important additional drug in combination therapy. However, despite the demonstrated efficacy of these mAb in the treatment of CLL, we still do not have a clear understanding of their mechanisms of action or the reasons for CLL cell resistance to mAb mediated cytotoxicity.
The potential cytotoxic mechanisms of mAb include complement dependent cytotoxicity (CDC), cell mediated cytotoxicity, and direct induction of cell death by apoptosis or autophagy. There is considerable in vitro data showing that ALM and RTX do not directly induce appreciable apoptosis in CLL cells [7–12]. In contrast there is extensive data showing that CDC is an important mechanism of action in CLL for ALM and OFA but not for RTX [9,10,13,14]. ALM, OFA, and RTX utilize a human IgG1 heavy chain constant region and are capable of activating antibody dependent cellular cytotoxicity (ADCC), and there is considerable data to support an important role for ADCC in the mechanism of action of these mAbs [12,15–21]. However, the functional importance of each of these mechanisms for these mAb in the treatment of CLL is still uncertain.
The rapid and extensive clearance of circulating CLL cells after initiation of ALM therapy in patients is likely to be substantially mediated by C3b-opsonization and CDC [22–24]. This cytotoxic reaction can be modeled in vitro and ALM in the presence of complement has previously been shown to rapidly kill 70%–80% of CLL cells in suspension culture [8,9]. It is likely that improving the efficacy of ALM-mediated CDC or increasing the level of CLL cell killing with an additional B cell targeting agent could improve clinical outcomes for patients with CLL. Several lines of evidence suggest that subpopulations of CLL cells can resist CDC mediated by a single mAb [9,10,25,26], and if the underlying mechanisms responsible for this resistance can be identified, it should be possible to develop more effective therapies. Potential mechanisms of CDC resistance include low mAb target expression, complement exhaustion, and increased activity or expression of complement regulatory proteins, which would result in decreased generation of membrane attack complexes (MAC) [11,27]. In addition, cell membranes can have increased intrinsic resistance to MAC mediated damage by mechanisms that include altered lipid synthesis [28].
The combination of complement activating mAb that target discrete cell-surface membrane proteins could potentially increase total CDC in a CLL cell population. One such combination is ALM (anti-CD52) and OFA (anti-CD20). Upon binding to B cells, OFA is very effective at activating complement and under comparable conditions promotes considerably more CDC than does RTX [13,14,29,30]. Thus OFA could be utilized to promote additional killing of CLL cells that are resistant to ALM induced CDC.
In this study we tested the hypothesis that in vitro OFA-mediated CDC increases the net killing of CLL cells targeted by ALM. Indeed, we found that OFA increases both complement activation (C3b and C5b-9 deposition) and CDC in CLL cells treated with ALM. However, in all patient samples we also discovered subpopulations of CLL cells that are resistant to CDC even after targeting with both mAbs. Identification of these resistant populations strongly suggests that small but potentially important subpopulations of CLL cells have intrinsic resistance to CDC.
Materials and Methods
Patients
The study was conducted at Mayo Clinic Rochester with the approval of the Institutional Review Board and according to the guidelines of the Declaration of Helsinki. We collected circulating CLL cells from 21 previously untreated patients with progressive CLL diagnosed using standard criteria [31,32]. Prognostic markers were evaluated using published methods [33–35]. Patient demographics and prognostic markers are summarized in Table I.
Table I.
Patient Demographics (n = 21)
| Gender | |
| Male | 16 (76.2%) |
|
| |
| Age (years) | |
| Median | 61 |
| Range | 50–80 |
|
| |
| Absolute lymphocyte count (× 109/L) | |
| Median | 78.3 |
| Range | 18.9–252.0 |
|
| |
| FISH * | |
| 17p13- | 1 (5%) |
| 11q22- | 1 (5%) |
| 12+ | 5 (24%) |
| No defect detected | 2 (10%) |
| 13q14- | 12 (57%) |
|
| |
| IGHV | |
| Unmutated (<2%) | 12 (57%) |
| Mutated (≥2%) | 9 (43%) |
|
| |
| ZAP-70 | |
| Positive (≥20%) | 12 (57%) |
| Negative (<20%) | 9 (43%) |
|
| |
| CD38 | |
| Positive (≥30%) | 5 (24%) |
| Negative (<30%) | 16 (76%) |
Described according to the hierarchical classification [48]
Specimen Collection
Peripheral blood mononuclear cells (PBMC) were isolated within 2 hours of blood collection from 20 mL of EDTA anticoagulated blood by density gradient centrifugation using Ficoll-Paque PLUS (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). The percentage of CD19+ CD5+ cells was determined by flow cytometry after staining with anti-CD19 and anti-CD5 antibodies (BD Biosciences, San Jose, CA) using a FACSCaliber (BD Biosciences). In 19 of 21 samples, the percentage of CLL cells (CD5+CD19+) was ≥90%. Two samples with <90% CLL cells required purification to ≥90% by negative selection (Human B Cell Enrichment Kit without CD43 Depletion, Stemcell Technologies, Vancouver, Canada). The median CLL cell content of studied samples was 95% (range 91 – 98%). Cell viability was assessed with trypan blue stain exclusion using the Countess Automated Cell Counter (Invitrogen, Carlsbad, CA) and determined to be ≥90% in all samples. Samples were then cryopreserved at a concentration of 20×106 cells/mL in freezing medium consisting of 10% dimethyl sulphoxide (Sigma-Aldrich, St. Louis, MO), 40% heat-inactivated fetal bovine serum (Sigma-Aldrich), and 50% RPMI 1640 medium (Gibco, Invitrogen Corporation, Grand Island, NY). Cell vials were first frozen overnight in a Cryo 1°C Freezing Container (Nalgene Labware, Thermo Fisher Scientific, Rochester, NY) at −80°C and then stored at −150°C. Prior to study, cells were rapidly thawed in a 37°C water bath, washed twice with 30mL of AIM-V medium (Invitrogen) and checked for viability by trypan blue staining. Samples with >90% cell viability were then incubated overnight under sterile conditions at 3×106 cells/mL in AIM-V medium at 37°C with 95% humidity and 5% CO2. Cells were then checked for viability by flow cytometry using annexin V and propidium iodide (PI)(BD Biosciences) as previously described to ensure viability >80% before being studied [9]. We performed CDC assays using RTX, OFA and ALM and measured CD20, CD52, and CD59 expression on samples from 3 patients before and after cryopreservation and thawing and found that storage of CLL cells using this method did not affect the results of CDC assays or the measured proteins expression levels (data not shown).
Quantification of CD20 and CD52 expression
CLL cells were treated with either mouse anti-human CD20-FITC antibodies (BD Biosciences) or mouse anti-human CD52-FITC (Pierce Antibodies, Thermo Fisher Scientific Inc., Rockford, IL). Target antigen expression was quantified by flow cytometry using molecules of equivalent soluble fluorochrome (MESF) with the Quantum FITC-5 MESF Premix kit (Bangs Laboratories, Fishers, IN) and QuickCal v2.3 software units as previously described [36].
CDC Assays
For these studies, the sources of complement were normal human serum (NHS)(Human Serum Complement, Sigma) and C5 deficient serum (C5-serum, Sigma). The complement levels of these reagents were assayed using standard clinical tests by the Mayo Clinic Protein Immunology Laboratory. Total complement activity of NHS was 55.7 U/mL and that of C5-serum was <3 U/mL (reference range 30–75 U/mL). The C3 level of NHS was 147 mg/dL and that of C5-serum was 141 mg/dL (reference range 75–175 mg/dL), and the C5 level of NHS was 25.5 mg/dL and that of C5-serum was <6 mg/dL (reference range 10.6–26.3 mg/dL). Rituximab (Genentech, South San Francisco, CA), ofatumumab (GlaxoSmithKline, Research Triangle Park, NC), and alemtuzumab (Genzyme, Cambridge, MA) were obtained from the Mayo Clinic pharmacy. CDC studies were performed as previously described [9]. In brief, 2×10°6 CLL cells in 1mL AIM-V medium were pretreated on ice for 30 minutes with 10μg/mL of each mAb either alone or in combination. These cell suspensions were then split and either received no serum, 10% (v/v) NHS, or 10% (v/v) C5-serum. The cells were then incubated for one hour at 37°C with 95% humidity and 5% CO2 and then washed twice with 3mL of AIM-V medium before analysis. Absolute viable cell counts were assayed using flow cytometry with BD Trucount beads (BD Biosciences) in a 1% BSA buffer [37] with PI staining to assess cell viability. Cells killed by CDC can either be lysed (i.e. disintegrated and no longer detected on flow cytometry) or remain intact with PI permeable membranes (intact dead cells). Cell lysis was determined for each sample by dividing the absolute cell count of the experimental specimens by that of the control specimen (10%NHS). Total CDC (% cytotoxicity) which measures the sum of lysed cells and intact dead cells was determined for each sample by dividing the absolute viable (PI negative) counts of the experimental specimens by that of the control specimen followed by multiplication by 100.
Analysis of CDC resistance
To analyze some of the factors that could contribute to the mechanisms of resistance of CLL cells that survived treatment with mAb and complement, 30×106 CLL cells were treated with 10μg/mL of ALM and 10%NHS. Surviving cells were then isolated by density gradient centrifugation and re-suspended in fresh AIM V medium. These surviving cells were then retreated with either 10%NHS (control) or 10%NHS with ALM, OFA, or ALM+OFA using the same methods as described above and evaluated for CDC. In samples from 3 patients, surviving cells after both initial and re-treatment were analyzed by flow cytometry for CD5+ and CD19+ expression as described above, and were shown to contain > 90% CLL cells.
Measurement of Complement Activation
We used flow cytometry to measure covalent deposition of C3 activation fragments or binding of C5b-9 to CLL cells. C3 fragment deposition was measured with the anti-C3b/iC3b FITC conjugated mAb 7C12 [38]. Binding of the terminal complement complex C5b-9 was measured by flow cytometry using a rabbit anti-human C5b-9 neoantigen antibody (Complement Technology, Inc., Tyler, TX) and a secondary Alexa fluor 488 goat anti-rabbit IgG antibody (H+L, Invitrogen). As noted above, CDC can cause complete disintegration of target cells (cell lysis) which is most likely to occur in the cells with the highest levels of complement activation. Therefore, to ensure that our studies measured C3 activation in the entire CLL population, these measurements were also conducted using C5-serum in which complement activation does not generate formation of the MAC responsible for cell lysis, but C3 fragment deposition occurs freely. For all of these experiments samples were incubated with the complement specific antibodies in the presence of 2mg/mL of purified mouse IgG (Lampire Biological Laboratories, Pipersville, PA) to decrease non-specific binding. Quantitation of binding was measured as delta median fluorescent intensity (dMFI) compared to the specimens not reacted with mAbs but simply treated with 10%NHS or 10%C5-serum only as appropriate.
Measurement of mAb Binding to CLL cells
We used flow cytometry and the anti-human Fc FITC conjugated mAb HB43 [38] to measure the binding of ALM, OFA or RTX to CLL cells. To evaluate mAb binding in all CLL cells these studies were performed on both cells treated with 10%NHS and those treated with 10% C5-serum.
Measurement of Expression of Complement Regulatory Proteins
The expression of CD59 and CD55 was measured by flow cytometry using anti-CD59 (BD Biosciences) and the anti-human CD55 FITC conjugated HD1A antibody [39].
Statistical Analysis
The Wilcoxon signed rank test was used to evaluate the relationship between paired values. Relationships between continuous variables were evaluated using Spearman's rank correlation coefficient. All tests were two-sided and statistical significance was defined as p<0.05.
Results
OFA increases ALM CDC
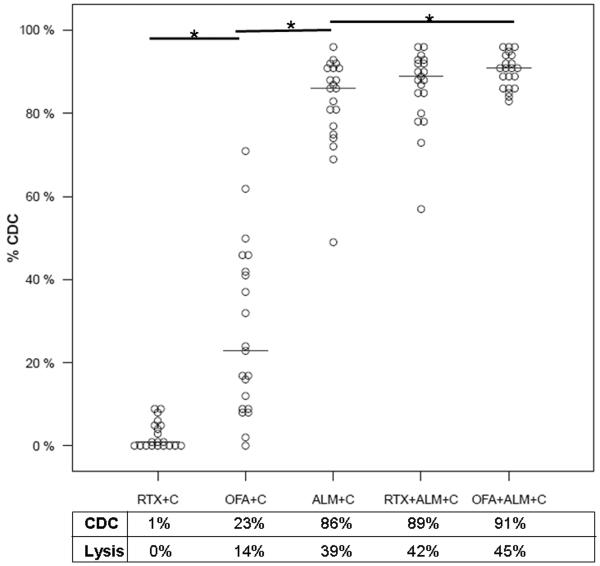
RTX induced low levels of CDC in CLL cells reacted in 10%NHS as a complement source (median 1%, range 0% – 9% p=0.002)(Figure 1). In contrast, under comparable conditions, OFA induced substantial CDC (median 23%, range 0%–71%, p<0.0001) and considerably higher levels of CDC were achieved by ALM (median 86%, range 49%–96%, p<0.0001)(Figure 1). CDC can cause cell lysis or dead cells can remain intact. In these studies, the median CLL cell lysis due to OFA mediated CDC was 14% (61% of total CDC) and 39% (45% of total CDC) for ALM mediated CDC. In general we found that approximately half of the cells killed by CDC were indeed lysed (Figure 1). The combination of ALM and OFA led to significantly higher median levels of CDC in CLL cells compared to those treated only with ALM (median 91%, range 83–96%, p = 0.01) (Figure 1). As apparent from the dot plot, the CLL cells with the lowest levels of ALM CDC had the largest increases in CDC with the combination of ALM and OFA.
Figure 1. Complement Dependent Cytotoxicity induced by Monoclonal Antibodies and Serum.
Complement dependent cytotoxicity (CDC) induced by monoclonal antibodies (mAb) was evaluated in vitro in CLL cells from 21 patients with untreated CLL using counting bead calibrated flow cytometry after propidium iodide staining. The results were normalized to measurements on CLL cells treated with 10% normal human serum (10%NHS) as a source of complement (C). These studies showed that CDC could result in cellular disintegration (lysis) or loss of cell membrane integrity without lysis (intact dead cells) and our results showed that these events occurred with similar frequency. Addition of rituximab and 10%NHS (RTX + C) promoted a low level of CDC compared to CLL cells treated with 10%NHS alone. However, ofatumumab and 10%NHS (OFA + C) did appreciably increase CDC (median 23%, p<0.0001) but with a wide range of responses (range 0% – 71%). Alemtuzumab and 10%NHS (ALM + C) induced considerably more CDC (median 86%, p<0.0001) with a narrower range of responses (49% - 96%). Addition of OFA to ALM +C caused significant additional increases in CDC (median 91%, p=0.01) with further narrowing of the range of response (83 – 96%) but this combination achieved > 95% cytotoxicity in only 3 patients. The median %CDC and %cell lysis for each mAb treatment is shown in the box below the dot plot and * indicate significant differences (p≤0.01).
1. mAb binding and CDC
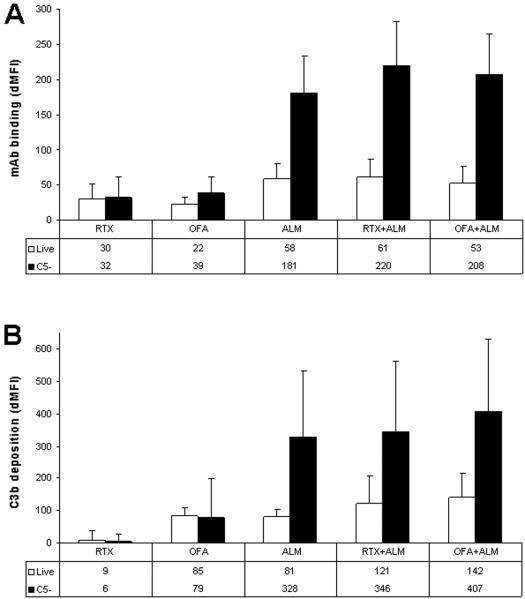
To investigate possible reasons for the difference in efficacy of ALM and OFA in mediating CDC we measured CD20 and CD52 expression by CLL cells and the ability of each mAb to bind to CLL cells and activate complement. The median expression of CD52 on CLL cells was significantly higher (6x) than that of CD20 (p<0.0001). We also found that there was considerably more binding of ALM to CLL cells compared to binding seen for either OFA or RTX (p<.0001) and that OFA binding was slightly higher than RTX (p=0.04)(Figure 2A).
Figure 2. mAb and C3b Binding.
CLL cells were treated with mAb and either 10%NHS (open bars) resulting in complement dependent cytotoxicity (CDC) in some cells or 10%C5-human serum (solid bars) resulting in complement activation but not CDC. Cells were then assayed by flow cytometry using counting beads, propidium iodide, anti-human Fc antibody, and anti-C3b antibody to measure binding of mAb and C3 activation fragments to live (propidium iodide negative) cells. The absolute counts were normalized using control experiments without the mAb. The bars represent the median values and error bars represent the standard deviation. A: For cells treated with C5-serum, rituximab (RTX) binding is slightly less than ofatumumab (OFA)(dMFI 32 vs 39, p = 0.04) which is considerably less than that of alemtuzumab (ALM)(dMFI 181, p<.0001). mAb binding is significantly higher in CLL cells treated with ALM, RTX + ALM, and OFA + ALM in C5-serum (black bars) compared to live cells reacted with NHS but not killed by CDC (open bars) (p<0.001). Compared to ALM alone, mAb binding in C5-serum was significantly higher for ALM + RTX (p<0.0001) and ALM + OFA (p=0.0049). B: C3b binding is significantly higher in CLL cells treated in C5-serum with ALM, RTX + ALM, and OFA + ALM (black bars) compared to the live cells (open bars) (p<0.0001). Compared to ALM alone, C3b binding in C5-serum was significantly higher for ALM + RTX (p=0.003) and ALM + OFA (p<0.0001).
We compared mAb binding to CLL cells in 10%NHS (contains all complement components and supports CDC) to mAb binding in 10%C5-serum (supports C3b activation but does not result in MAC formation and CDC) (Figure 2A). Live (PI negative) CLL cells treated with OFA, ALM, RTX + ALM, and OFA + ALM in 10%C5-serum has significantly higher levels of mAb binding compared to those reacted in 10%NHS (p<0.001 for each comparison)(Figure 2A). The higher levels of binding of the mAbs (and of C3b, see below) to cells reacted in C5-serum compared to intact NHS most likely reflects the killing of CLL cells with the highest levels of mAb ligation and thus complement activation in intact serum. We are currently investigating this question and will report on it separately.
2. C3b deposition and CDC
C3b binding was considerably higher in CLL cells treated with ALM, RTX + ALM, and OFA + ALM in C5-serum compared to binding observed for live (CDC resistant) cells following treatment with 10%NHS (p<0.0001)(Figure 2B). These results for cells treated with ALM suggest that higher levels of C3b deposition are associated with increased amounts of CDC. In contrast, C3b binding to CLL cells treated with OFA in 10%C5-serum was similar to C3b binding to live cells following treatment with 10%NHS (p=0.55)(Figure 2B). This finding suggests that the susceptibility of CLL cells to OFA mediated CDC is less dependent on the aggregate level of C3b activation and that there could be differences in the mechanisms by which OFA and ALM induce CDC. This is currently under investigation and will be reported separately.
3. Binding of C5b-9 to mAb opsonized CLL cells
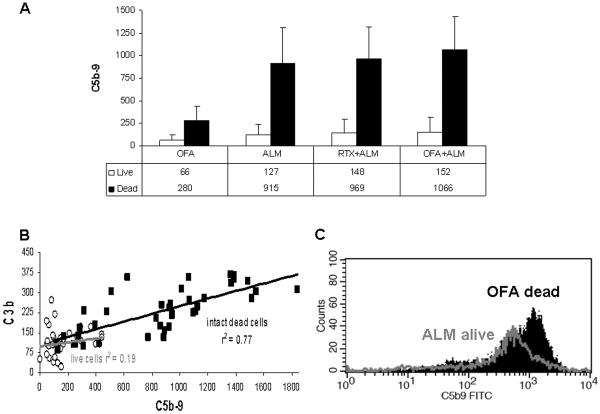
ALM, OFA and RTX can activate the classical complement pathway with sequential generation of C3b and C5b-9 resulting in formation of the MAC. To determine the relationship between C3b binding and generation of the MAC we measured levels of C5b-9 in CLL cells from 10 patients treated with mAb and 10%NHS. As illustrated in Figure 3A, in all cases median C5b-9 levels were considerably higher in intact dead cells (PI positive) compared to live cells (PI negative)(p=0.002). In addition, considerably more C5b-9 binding was detected in cells reacted with ALM compared to cells reacted with OFA (live and intact dead cells p≤0.006). Addition of RTX or OFA to ALM modestly increased C5b-9 levels in intact dead cells (p=0.01 and p=0.02 respectively) but not in live cells (p>0.2). Figure 3B indicates that there was a significant correlation between the levels of deposited C3b (which includes data from Figure 1) and cell-associated C5b-9 for the intact dead CLL cells (squares, r2=0.77, p<.0001). This was also found for each individual population of dead cells: OFA (p=0.01); ALM (p<0.0001); ALM + RTX (p=0.02); and ALM + OFA (p=0.02). On the other hand, no correlation was evident for live cells (open circles, r2=0.19, p=0.23) for any population (Figure 3B). Moreover, many of the live cells had as much cell-associated C3b or C5b-9 as did the dead cells, but obviously this level of complement attack was not adequate to kill the cells. This suggests that there is a subpopulation of CLL cells that is very sensitive to mAb induced CDC (killed by lower levels of C5b-9 mediated by binding of OFA) and another subpopulation that is more resistant to the effects of complement activation (not killed by higher levels of C5b-9 generated by ALM). An individual patient with CLL could thus have appreciable variation in the sensitivity of their CLL cells to activated complement, with distinct subpopulations of cells which are susceptible and resistant to MAC activity (Figure 3C).
Figure 3. C3b and C5b-9 Expression and Complement Dependent Cytotoxicity.
C5b-9 levels were measured on CLL cells from a subset of 10 patients including all 5 patients with < 75% CDC on initial treatment with mAb and 10% normal human serum (NHS). A: C5b-9 binding was measured by flow cytometry in viable (propidium iodide (PI) negative) and intact dead (PI positive) CLL cells. The median C5b-9 binding (dMFI) for viable cells (open bars) and intact dead (black bars) is shown with error bars representing the standard deviation. C5b-9 binding was significantly higher in intact dead cells compared to viable cells for all cases (p=0.002). The level of binding of C5b-9 was higher in cells treated with ALM compared to OFA (live and intact dead cells p≤0.006). Addition of OFA (p=0.02) and RTX (p=0.01) to ALM significantly increased C5b-9 binding in intact dead cells but not viable cells (p>0.1). B: The binding of C3b and C5b-9 was compared in viable (open circles) and intact dead (black squares) CLL cells treated, in 10% NHS, with OFA, ALM, ALM + RTX, and ALM + OFA. There was a significant linear relationship between binding levels of C3b and C5b-9 in intact dead cells (black squares, r2=0.77, p<.0001) but not live cells (open circles r2=0.19, p=0.23). There was also considerable overlap in C5b-9 levels between viable and intact dead cells for both of these measurements. C: CLL cells from a representative patient were treated with 10%NHS and either ALM or OFA. This histogram shows C5b-9 binding to viable cells treated with ALM (grey) and intact dead cells (black) with OFA.
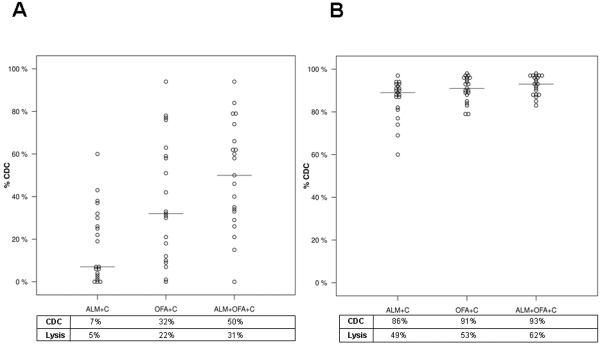
4. OFA increases ALM CDC
To test the efficacy of sequential therapy with ALM followed by OFA, CLL cells surviving initial treatment with ALM and 10%NHS were retreated with ALM, OFA, or ALM + OFA, and 10%NHS. This resulted in a significantly higher median CDC (32%) for OFA compared to ALM (7%)(p=0.01)(Figure 4A). The cumulative CDC of concomitant and sequential (91% in both Figures 1 and 4B) therapy with ALM and OFA was thus similar. Our data shows that addition of OFA to ALM increases CDC of CLL cells in vitro. However it is important to note that approximately 10% of the CLL cells remain resistant to combination therapy indicating that this combination mAb therapy achieves only 1 log order cytotoxicity in vitro. This CDC resistant CLL cell population could limit the clinical efficacy of combination mAb therapy.
Figure 4. Retreatment of CLL Cells Surviving ALM CDC.
CLL cells were first treated with ALM and 10%NHS and the surviving viable intact cells were then isolated by density centrifugation. These cells were subsequently reacted in 10%NHS with ALM, OFA, or ALM + OFA and viable absolute cell counts were measured and normalized to the counts of cells treated with only 10%NHS. A: Retreatment of CLL cells with ALM and 10%NHS resulted in little additional cytotoxicity in most patients (median 7%)(p > 0.1) although some samples did show more CDC. The median %CDC was significantly higher for cells treated with OFA (32%)(p=0.01) or the combination of ALM and OFA (50%)(p=0.01). B: The cumulative cytotoxicity achieved by sequential treatment of CLL cells with ALM and 10%NHS followed by isolation of viable cells and treatment of these cells with ALM, OFA or ALM + OFA with 10%NHS. These data show that higher CDC was achieved using OFA than using ALM alone (p=0.01) and that the highest CDC was achieved with ALM + OFA (p=0.01). The median %CDC (represented by the horizontal bar in dot plots) and %cell lysis for each mAb treatment is shown in the box below the dot plots.
Mechanisms of resistance of CLL cells to mAb CDC
To examine possible reasons why a subpopulation of CLL cells is resistant to mAb mediated CDC, we tested for limiting factors (availability of complement and mAb), the role of complement regulatory proteins (CRP), and the intrinsic ability of the CLL cells to resist the cytotoxic effects of activated complement.
1. mAb and complement availability are not limiting factors for CDC
Re-treatment of viable CLL cells, that had survived initial treatment with ALM and 10%NHS, with fresh 10%NHS resulted in no significant increase in CDC (median 1%, range 0%-6%, p=0.0005). Re-treatment of these viable cells with ALM and 10%NHS, resulted in only a modest additional mAb and C3b binding and cytotoxicity (median 7% CDC, range 0%–60% Figure 4A) which was significantly lower than the CDC (median 86%, range 49%–96%) achieved with initial treatment (p<.0001)(Figure 1). These data suggest that ALM and complement are not limiting factors for CDC in this experimental model.
2. High levels of C3b deposition are necessary but not sufficient for CDC
A subpopulation of CLL cells could be resistant to CDC because of lower levels of complement activation. As reported above, we compared the binding of C3b and C5b-9 to viable and intact dead cells after treatment with mAb and 10%NHS (Figure 3B). These data show that higher levels of C3b binding were associated with higher levels of C5b-9 binding in the cells that were killed but intact and thus amenable to analysis. However, as noted above, there was considerable overlap between the levels of C3b binding between live (open circles) and intact dead cells (filled squares).
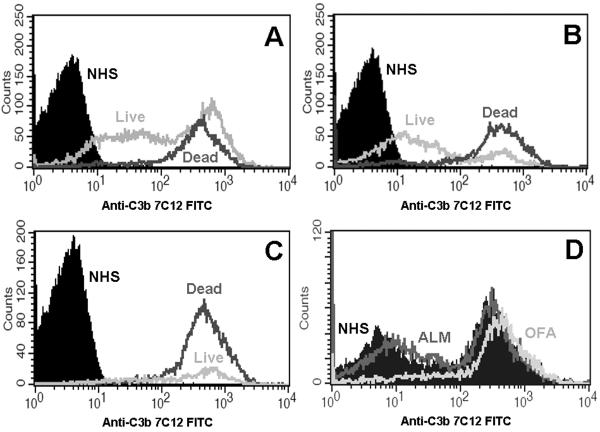
To further evaluate the importance of C3b deposition, we next examined the detailed patterns of C3b deposition in the CLL cells. As shown in data from a representative patient, we found that C3b binding following reaction with OFA (Figure 5A, 10% CDC) or ALM (Figure 5B, 50% CDC) in 10%NHS was bimodal for cells resistant to CDC (live) and unimodal for intact dead cells killed by CDC. Moreover, for both mAb treatments there was a sizable subpopulation of live cells with C3b binding equal to that found on intact dead cells. The CLL cells from this patient were then either treated with ALM +OFA concomitantly in a cocktail (87% CDC), or sequentially (total CDC of 80%). Concomitant treatment resulted in a marked increase in C3b deposition in all cells and the small subpopulation of live cells had the same level of cell-associated C3b as the intact dead cells (Figure 5C). When live CLL cells that survived treatment with ALM and 10%NHS were purified, resuspended in fresh medium, and then treated again with either OFA or ALM and 10%NHS, examination of the live cells surviving the second treatment shows that OFA promoted more C3b deposition than ALM but that these cells did not undergo CDC (Figure 5D).
Figure 5. Comparison of C3b Activation in Live and Intact Dead CLL Cells.
These data from a representative patient sample compare C3b binding in live and intact dead CLL cells treated with mAb and 10%NHS. A: CLL cells were treated with OFA and 10%NHS resulting in 10% CDC. C3b binding levels were high on all intact dead cells. In cells surviving CDC, there was a bimodal distribution of C3b binding with one mode having binding equivalent to that observed in intact dead cells and the other having lower levels of C3b binding. B: CLL cells were treated with ALM and 10%NHS resulting in 50% CDC. All the intact dead cells had high levels of C3b binding and there was bimodal binding of C3b in cells that survived CDC. C: CLL cells were treated with ALM + OFA and 10%NHS resulting in 87% CDC. The small number of CLL cells that survived treatment had high levels of C3b binding which were equivalent to that of intact dead cells. D: CLL cells were treated with ALM and 10%NHS and the live cells isolated by density centrifugation. These cells were then treated again with either ALM and 10%NHS (16% CDC) or OFA and 10%NHS (59% CDC) and the live cells evaluated for C3b binding. This figure shows bimodal binding of C3b in live cells treated with 10%NHS and ALM and 10%NHS. Cells treated with OFA and 10%NHS had higher levels of C3b binding which was unimodal. However, despite the high levels of C3b binding in many of these CLL cells, they did not undergo CDC.
These data show that high levels of C3b deposition are required for CDC but that not all cells opsonized with large amounts of C3b are killed by CDC. We have previously shown that there is no increase in the rate of cell death in cells surviving mAb mediated CDC compared to control cells at 24 hours after treatment [9] and thus conclude that a subpopulation of CLL cells is resistant to CDC despite effective activation of C3b.
3. Individual patients have a subpopulation of CLL cells that is resistant to activated complement
CLL cells that capture large amounts of activated C3b could fail to undergo CDC because of inadequate generation of the MAC. To test for inadequate MAC generation we measured C5b-9 binding to CLL cells from a subpopulation of 10 patients in this study including all 5 patients that had < 75% ALM CDC. As reported above, these studies showed a significant correlation between C3b deposition and C5b-9 incorporation in intact dead cells but not viable cells (Figure 3B). However, a subpopulation of viable CLL cells had high levels of C5b-9 binding which suggests an intrinsic cellular resistance to the effects of MAC formation (Figure 3C). These data suggest that both inhibition of MAC formation and resistance to the effects of the MAC could be important in the resistance of CLL cells to CDC.
4. The role of CRP in CLL cell resistance to CDC
Our finding that for almost all patients there is a subpopulation of cells with high levels of C3b deposition but low levels of binding of C5b-9 (Figure 3B) suggests that higher levels of activity of CPR in these patients could contribute to resistance to mAb induced CDC. CD46 has been previously shown to have uniform low expression in CLL cells [10] and expression of this CRP was thus not examined in this study. CD55 inhibits the generation and activity of C3 and C5 convertases [40] and CD59 prevents assembly of the terminal complement complex [41]. We measured CD59 (n=21) and CD55 (n=6) expression by the CLL cells and did not find any significant differences in levels of expression in viable and intact dead cells after treatment with mAb and 10%NHS and resistance to CDC (data not shown). Specifically, CLL cells resistant to CDC did not have increased levels of expression of CD59 or CD55. In addition, we found no correlation between the levels of expression of CD59 in untreated CLL cells from the 21 patients and their subsequent in vitro CDC (r2=0.01, p=0.98). However, we did not test the activity of CD55 or CD59 in these cells and were thus not able to determine the reason for failure of C3b deposition to promote robust MAC generation adequate for CDC on subpopulations of cells.
Discussion
We have shown that OFA increases ALM CDC in vitro which provides pre-clinical data supporting a clinical trial testing the efficacy and safety of the combination of ALM and OFA in the treatment of CLL. Our study has also shown that for almost all of the patients, there is a subpopulation of CLL cells that are resistant to mAb mediated CDC in vitro even after two consecutive treatments. We have confirmed the findings of previously published studies which showed that in a subpopulation of CDC-resistant CLL cells with high levels of C3b deposition, there was inadequate generation of the MAC. This phenomenon could not be explained based on differences in levels of expression of the CRP CD55 and CD59 in the CDC resistant and susceptible populations [8]. In addition, we describe the novel finding that within the subpopulation of CDC resistant CLL cells there are mAb-opsonized cells that have indeed activated complement, because they have large amounts of both deposited C3b and C5b-9. These data show that resistance of CLL cells to CDC mediated by mAb is multifactorial and suggest several potential opportunities for overcoming this resistance.
Therapy of CLL with the combination of ALM and RTX is effective with response rates that are higher than those reported for ALM monotherapy [24,42–44]. However, complete responses in patients with relapsed/refractory disease are uncommon and the duration of response is limited [42]. Clearly better combination therapies are required. Because OFA has single agent activity in patients with relapsed/refractory CLL monotherapy [6], there is a rationale for combination therapy with ALM and OFA. Our in vitro data shows that OFA CDC is significantly higher than that of RTX and that OFA improves ALM CDC when used together and sequentially. This is important additional justification for developing a clinical trial to determine the efficacy and safety of OFA and ALM combination therapy.
In this study we used an in vitro model of CDC to compare two anti-CD20 mAb (RTX and OFA) and an anti-CD52 mAb (ALM), and as previously reported, OFA induced considerably more CDC than RTX [13,14]. However, OFA induced less CDC than ALM which correlates well with the much lower level of CD20 expression compared to that of CD52. These data support the importance of antigen density in influencing the efficacy of complement activation by mAb.
An important finding was that a subpopulation of CLL cells that survived treatment with ALM and 10%NHS retained sensitivity to OFA CDC. In these ALM CDC “resistant” cells, OFA had lower median levels of cell binding but higher levels of C3b deposition and CDC compared to cells reacted with ALM. This observation suggests there may be differences in the mechanisms of complement activation by OFA and ALM. Potential explanations include spatial separation of CD20 and CD52 on the cell membrane and differences in movement of these molecules in the cell membrane after mAb ligation. Additional investigations will be required in the future and could provide important insights into the mechanisms of action of ALM and OFA.
Our studies of the level of binding of C3b and C5b-9 to CLL cells after treatment with mAb and 10%NHS showed that ALM was the most effective antibody at activating complement and levels of C3b deposition correlated with levels of binding of C5b-9 in intact CLL cells killed by CDC. However, levels of C3b and C5b-9 did not correlate well in surviving cells suggesting there may be several mechanisms by which the cells resist CDC. To further investigate this question we measured the expression of the CRP CD55 and CD59 by CLL cells but did not find increased levels of expression of these proteins in cells that were not killed by CDC. This finding is similar to that previously reported where, despite the failure to show differences in levels of expression of CRP in CDC sensitive and resistant CLL cells, inhibition of CD59 and CD55 activity did increase CDC [10,45].
Resistance to the cytotoxic effects of the MAC has been previously demonstrated for several types of cells [28]. This present study has demonstrated for the first time, that a subpopulation of CLL cells survive the effects of complement activation despite levels of membrane bound C5b-9 that are equal to or higher than on cells that are killed by CDC (Fig. 3B–C). Cells that are intrinsically resistant to MAC cytotoxicity have previously been shown to use multiple defense mechanisms [46]. Our novel identification of an intrinsically CDC resistant subpopulation of CLL cells provides an opportunity to identify the mechanisms of this resistance so as to be able to develop combination regimens to overcome resistance and improve treatment efficacy.
CDC resistance in CLL cells could also be overcome by interventions to increase cellular cytotoxicity. One possible intervention is to target complement components, selectively deposited on mAb opsonized cells, such as iC3b. Binding of this complement fragment to complement receptor 3 (CR3) expressed by neutrophils and macrophages can result in complement dependent cellular cytotoxicity that can be enhanced by soluble CR3-specific polysaccharides such as beta-glucan [47]. Our finding that most CDC resistant cells express high C3b/iC3b levels supports further investigation of these therapeutic modalities.
This study used peripheral blood CLL cells from previously untreated patients with progressive disease. This is an important target population for therapy according to current clinical criteria and thus the study does use specimens from a relevant and appropriate cohort. However, the study included only one patient with 17p13-, a genetic defect associated with very-high risk CLL. The finding that OFA increases ALM induced CDC in CLL cells will thus need to be further tested in more samples from patients with very-high risk disease (p53 dysfunction and purine analogue refractory disease) in future studies. The CDC assay takes into account the important observation that a considerable number of CLL cells treated in vitro with mAb and complement undergo lysis and will thus not be detectible using assays that measure only the percentage of viable cells. We also ensured that the subpopulations of resistant cells were indeed CLL cells. Our in vitro experiments thus used specimens from appropriate patients with assays that provide accurate measurements of CDC. This CDC model used CLL cells at concentrations that are considerably lower than those occurring in the circulation of patients with CLL. This ensured that there were adequate cells to do all the experiments and that mAb and complement availability were optimized for detection of CLL cell sensitivity and resistance to CDC. However, the limitation of this model was that we were not able to determine the role of potentially important factors leading to resistance to mAb cytotoxicity including low levels of mAb and complement and factors affecting ADCC. Despite these limitations, many of which are intrinsic to in vitro testing, we believe that our study has provided important novel data on which to base future studies aimed at improving treatment of patients with CLL. Our ongoing correlative studies in patients on clinical trials for treatment using mAb are designed to overcome these limitations.
In conclusion, we have shown that OFA increases CDC of CLL cells when added to ALM but that this combination is unable to kill a small subpopulation of cells that appear to be intrinsically resistant to activated complement. This combination therapy is unlikely to achieve the sequential 2–3 log order reduction of tumor burden per cycle of treatment that could lead to eradication of the CLL clone and result in cure of the disease. However, our study suggests several potentially important mechanisms of resistance that could be targeted to improve the effectiveness of treatment.
Acknowledgments
This work was supported in part by a grant from the National Institutes of Health, USA (University of Iowa/Mayo Clinic Lymphoma SPORE CA097274), and the University of Iowa/Mayo Clinic Lymphoma SPORE Foundation with funds provided in memory of Dr. Ruth Westrick Connolly and by CLL Topics.
References
- 1.Stilgenbauer S, Zenz T. Understanding and managing ultra high-risk chronic lymphocytic leukemia. Hematology Am Soc Hematol Educ Program. 2010:481–488. doi: 10.1182/asheducation-2010.1.481. [DOI] [PubMed] [Google Scholar]
- 2.Keating MJ, O'brien S, Albitar M, Lerner S, Plunkett W, Giles F, Andreeff M, Cortes J, Faderl S, Thomas D, et al. Early results of a chemoimmunotherapy regimen of fludarabine, cyclophosphamide, and rituximab as initial therapy for chronic lymphocytic leukemia. J Clin Oncol. 2005;23:4079–4088. doi: 10.1200/JCO.2005.12.051. [DOI] [PubMed] [Google Scholar]
- 3.Kay NE, Geyer SM, Call TG, Shanafelt TD, Zent CS, Jelinek DF, Tschumper R, Bone ND, Dewald GW, Lin TS, et al. Combination chemoimmunotherapy with pentostatin, cyclophosphamide, and rituximab shows significant clinical activity with low accompanying toxicity in previously untreated B chronic lymphocytic leukemia. Blood. 2007;109:405–11. doi: 10.1182/blood-2006-07-033274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrd JC, Rai K, Peterson BL, Appelbaum FR, Morrison VA, Kolitz JE, Shepherd L, Hines JD, Schiffer CA, Larson RA. Addition of rituximab to fludarabine may prolong progression-free survival and overall survival in patients with previously untreated chronic lymphocytic leukemia: an updated retrospective comparative analysis of CALGB 9712 and CALGB 9011. Blood. 2005;105:49–53. doi: 10.1182/blood-2004-03-0796. [DOI] [PubMed] [Google Scholar]
- 5.Hallek M, Fischer K, Fingerle-Rowson G, Fink AM, Busch R, Mayer J, Hensel M, Hopfinger G, Hess G, von Grunhagen U, et al. Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: a randomised, open-label, phase 3 trial. Lancet. 2010;376:1164–74. doi: 10.1016/S0140-6736(10)61381-5. [DOI] [PubMed] [Google Scholar]
- 6.Wierda WG, Kipps TJ, Mayer J, Stilgenbauer S, Williams CD, Hellmann A, Robak T, Furman RR, Hillmen P, Trneny M, et al. Ofatumumab as single-agent CD20 immunotherapy in fludarabine-refractory chronic lymphocytic leukemia. J Clin Oncol. 2010;28:1749–55. doi: 10.1200/JCO.2009.25.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zent CS, Chen JB, Kurten RC, Kaushal GP, Lacy HM, Schichman SA. Alemtuzumab (CAMPATH 1H) does not kill chronic lymphocytic leukemia cells in serum free medium. Leuk Res. 2004;28:495–507. doi: 10.1016/j.leukres.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Golay J, Manganini M, Rambaldi A, Introna M. Effect of alemtuzumab on neoplastic B cells. Haematologica. 2004;89:1476–1483. [PubMed] [Google Scholar]
- 9.Zent CS, Secreto CR, Laplant BR, Bone ND, Call TG, Shanafelt TD, Jelinek DF, Tschumper RC, Kay NE. Direct and complement dependent cytotoxicity in CLL cells from patients with high-risk early-intermediate stage chronic lymphocytic leukemia (CLL) treated with alemtuzumab and rituximab. Leuk Res. 2008;32:1849–1856. doi: 10.1016/j.leukres.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golay J, Lazzari M, Facchinetti V, Bernasconi S, Borleri G, Barbui T, Rambaldi A, Introna M. CD20 levels determine the in vitro susceptibility to rituximab and complement of B-cell chronic lymphocytic leukemia: further regulation by CD55 and CD59. Blood. 2001;98:3383–3389. doi: 10.1182/blood.v98.12.3383. [DOI] [PubMed] [Google Scholar]
- 11.Taylor RP, Lindorfer MA. Immunotherapeutic mechanisms of anti-CD20 monoclonal antibodies. Curr Opin Immunol. 2008;20:444–9. doi: 10.1016/j.coi.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glennie MJ, French RR, Cragg MS, Taylor RP. Mechanisms of killing by anti-CD20 monoclonal antibodies. Mol Immunol. 2007;44:3823–37. doi: 10.1016/j.molimm.2007.06.151. [DOI] [PubMed] [Google Scholar]
- 13.Teeling JL, French RR, Cragg MS, van den Brakel J, Pluyter M, Huang H, Chan C, Parren PW, Hack CE, Dechant M, et al. Characterization of new human CD20 monoclonal antibodies with potent cytolytic activity against non-Hodgkin lymphomas. Blood. 2004;104:1793–800. doi: 10.1182/blood-2004-01-0039. [DOI] [PubMed] [Google Scholar]
- 14.Pawluczkowycz AW, Beurskens FJ, Beum PV, Lindorfer MA, van de Winkel JG, Parren PW, Taylor RP. Binding of submaximal C1q promotes complement-dependent cytotoxicity (CDC) of B cells opsonized with anti-CD20 mAbs ofatumumab (OFA) or rituximab (RTX): considerably higher levels of CDC are induced by OFA than by RTX. Journal of immunology. 2009;183:749–58. doi: 10.4049/jimmunol.0900632. [DOI] [PubMed] [Google Scholar]
- 15.Clynes R, Towers T, Presta L, Ravetch J. Inhibitory Fc receptors modulate in vivo cytoxicity against tumor targets. Nat Med. 2000;6:443–446. doi: 10.1038/74704. [DOI] [PubMed] [Google Scholar]
- 16.Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, Watier H. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood. 2002;99:754–8. doi: 10.1182/blood.v99.3.754. [DOI] [PubMed] [Google Scholar]
- 17.Bowles JA, Weiner GJ. CD16 polymorphisms and NK activation induced by monoclonal antibody-coated target cells. J Immunol Methods. 2005;304:88–99. doi: 10.1016/j.jim.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 18.Golay J, Cortiana C, Manganini M, Cazzaniga G, Salvi A, Spinelli O, Bassan R, Barbui T, Biondi A, Rambaldi A, et al. The sensitivity of acute lymphoblastic leukemia cells carrying the t(12;21) translocation to campath-1H-mediated cell lysis. Haematologica. 2006;91:322–30. [PubMed] [Google Scholar]
- 19.Bleeker WK, Munk ME, Mackus WJ, van den Brakel JH, Pluyter M, Glennie MJ, van de Winkel JG, Parren PW. Estimation of dose requirements for sustained in vivo activity of a therapeutic human anti-CD20 antibody. British journal of haematology. 2008;140:303–12. doi: 10.1111/j.1365-2141.2007.06916.x. [DOI] [PubMed] [Google Scholar]
- 20.Siders WM, Shields J, Garron C, Hu Y, Boutin P, Shankara S, Weber W, Roberts B, Kaplan JM. Involvement of neutrophils and natural killer cells in the anti-tumor activity of alemtuzumab in xenograft tumor models. Leuk Lymphoma. 2010;51:1293–304. doi: 10.3109/10428191003777963. [DOI] [PubMed] [Google Scholar]
- 21.Kohrt HE, Houot R, Goldstein MJ, Weiskopf K, Alizadeh AA, Brody J, Muller A, Pachynski R, Czerwinski D, Coutre S, et al. CD137 stimulation enhances the antilymphoma activity of anti-CD20 antibodies. Blood. 2011;117:2423–32. doi: 10.1182/blood-2010-08-301945. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Lundin J, Osterborg A, Brittinger G, Crowther D, Dombret H, Engert A, Epenetos A, Gisselbrecht C, Huhn D, Jaeger U, et al. CAMPATH-1H monoclonal antibody in therapy for previously treated low-grade non-Hodgkin's lymphomas: a phase II multicenter study. European Study Group of CAMPATH-1H Treatment in Low-Grade Non-Hodgkin's Lymphoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1998;16:3257–63. doi: 10.1200/JCO.1998.16.10.3257. [DOI] [PubMed] [Google Scholar]
- 23.Rawstron AC, Kennedy B, Moreton P, Dickinson AJ, Cullen MJ, Richards SJ, Jack AS, Hillmen P. Early prediction of outcome and response to alemtuzumab therapy in chronic lymphocytic leukemia. Blood. 2004;103:2027–31. doi: 10.1182/blood-2002-10-3270. [DOI] [PubMed] [Google Scholar]
- 24.Zent CS, Call TG, Shanafelt TD, Tschumper RT, Jelinek DF, Bowen DA, Secreto CR, LaPlant BR, Kabat BF, Kay NE. Early treatment of high risk chronic lymphocytic leukemia with alemtuzumab and rituximab. Cancer. 2008;113:2110–2118. doi: 10.1002/cncr.23824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beum PV, Lindorfer MA, Taylor RP. Within peripheral blood mononuclear cells, antibody-dependent cellular cytotoxicity of rituximab-opsonized Daudi cells is promoted by NK cells and inhibited by monocytes due to shaving. J Immunol. 2008;181:2916–24. doi: 10.4049/jimmunol.181.4.2916. [DOI] [PubMed] [Google Scholar]
- 26.Kennedy AD, Beum PV, Solga MD, DiLillo DJ, Lindorfer MA, Hess CE, Densmore JJ, Williams ME, Taylor RP. Rituximab infusion promotes rapid complement depletion and acute CD20 loss in chronic lymphocytic leukemia. J Immunol. 2004;172:3280–3288. doi: 10.4049/jimmunol.172.5.3280. [DOI] [PubMed] [Google Scholar]
- 27.Bohana-Kashtan O, Ziporen L, Donin N, Kraus S, Fishelson Z. Cell signals transduced by complement. Mol Immunol. 2004;41:583–97. doi: 10.1016/j.molimm.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Black SM, Schott ME, Batdorf BH, Benson BA, Rutherford MS, Levay-Young BK, Dalmasso AP. IL-4 induces protection of vascular endothelial cells against killing by complement and melittin through lipid biosynthesis. European journal of immunology. 2010;40:803–12. doi: 10.1002/eji.200939488. [DOI] [PubMed] [Google Scholar]
- 29.Beum PV, Lindorfer MA, Beurskens F, Stukenberg PT, Lokhorst HM, Pawluczkowycz AW, Parren PW, van de Winkel JG, Taylor RP. Complement activation on B lymphocytes opsonized with rituximab or ofatumumab produces substantial changes in membrane structure preceding cell lysis. J Immunol. 2008;181:822–32. doi: 10.4049/jimmunol.181.1.822. [DOI] [PubMed] [Google Scholar]
- 30.Teeling JL, Mackus WJ, Wiegman LJ, van den Brakel JH, Beers SA, French RR, van Meerten T, Ebeling S, Vink T, Slootstra JW, et al. The biological activity of human CD20 monoclonal antibodies is linked to unique epitopes on CD20. J Immunol. 2006;177:362–71. doi: 10.4049/jimmunol.177.1.362. [DOI] [PubMed] [Google Scholar]
- 31.Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, Hillmen P, Keating MJ, Montserrat E, Rai KR, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia (IWCLL) updating the National Cancer Institute-Working Group (NCI-WG) 1996 guidelines. Blood. 2008;111:5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morice WG, Kurtin PJ, Hodnefield JM, Shanafelt TD, Hoyer JD, Remstein ED, Hanson CA. Predictive value of blood and bone marrow flow cytometry in B-cell lymphoma classification: comparative analysis of flow cytometry and tissue biopsy in 252 patients. Mayo Clin Proc. 2008;83:776–85. doi: 10.4065/83.7.776. [DOI] [PubMed] [Google Scholar]
- 33.Dewald GW, Brockman SR, Paternoster SF, Bone ND, O'Fallon JR, Allmer C, James CD, Jelinek DF, Tschumper RC, Hanson CA, et al. Chromosome anomalies detected by interphase fluorescence in situ hybridization: correlation with significant biological features of B-cell chronic lymphocytic leukaemia. Br J Haematol. 2003;121:287–295. doi: 10.1046/j.1365-2141.2003.04265.x. [DOI] [PubMed] [Google Scholar]
- 34.Jelinek DF, Tschumper RC, Geyer SM, Bone ND, Dewald GW, Hanson CA, Stenson MJ, Witzig TE, Tefferi A, Kay NE. Analysis of clonal B-cell CD38 and immunoglobulin variable region sequence status in relation to clinical outcome for B-chronic lymphocytic leukaemia. Br J Haematol. 2001;115:854–861. doi: 10.1046/j.1365-2141.2001.03149.x. [DOI] [PubMed] [Google Scholar]
- 35.Rassenti LZ, Huynh L, Toy TL, Chen L, Keating MJ, Gribben JG, Neuberg DS, Flinn IW, Rai KR, Byrd JC, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med. 2004;351:893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 36.Rossmann ED, Lenkei R, Lundin J, Mellstedt H, Osterborg A. Performance of calibration standards for antigen quantitation with flow cytometry in chronic lymphocytic leukemia. Cytometry B Clin Cytom. 2007;72:450–7. doi: 10.1002/cyto.b.20359. [DOI] [PubMed] [Google Scholar]
- 37.Brando B, Gohde W, Jr., Scarpati B, D'Avanzo G. The “vanishing counting bead” phenomenon: effect on absolute CD34+ cell counting in phosphate-buffered saline-diluted leukapheresis samples. Cytometry. 2001;43:154–60. doi: 10.1002/1097-0320(20010201)43:2<154::aid-cyto1031>3.3.co;2-u. [DOI] [PubMed] [Google Scholar]
- 38.Edberg JC, Tosic L, Wright EL, Sutherland WM, Taylor RP. Quantitative analyses of the relationship between C3 consumption, C3b capture, and immune adherence of complement-fixing antibody/DNA immune complexes. J Immunol. 1988;141:4258–65. [PubMed] [Google Scholar]
- 39.Harris CL, Lublin DM, Morgan BP. Efficient generation of monoclonal antibodies for specific protein domains using recombinant immunoglobulin fusion proteins: pitfalls and solutions. J Immunol Methods. 2002;268:245–58. doi: 10.1016/s0022-1759(02)00207-7. [DOI] [PubMed] [Google Scholar]
- 40.Macor P, Tedesco F. Complement as effector system in cancer immunotherapy. Immunol Lett. 2007;111:6–13. doi: 10.1016/j.imlet.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 41.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–97. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faderl S, Thomas DA, O'Brien S, Garcia-Manero G, Kantarjian HM, Giles FJ, Koller C, Ferrajoli A, Verstovsek S, Pro B, et al. Experience with alemtuzumab plus rituximab in patients with relapsed and refractory lymphoid malignancies. Blood. 2003;101:3413–3415. doi: 10.1182/blood-2002-07-1952. [DOI] [PubMed] [Google Scholar]
- 43.Nabhan C, Patton D, Gordon L, Riley M, Kuzel T, Tallman M, Rosen S. A Pilot Trial of Rituximab and Alemtuzumab Combination Therapy in Patients with Relapsed and/or Refractory Chronic Lymphocytic Leukemia (CLL) Leuk Lymphoma. 2004;45:2269–2273. doi: 10.1080/10428190412331286096. [DOI] [PubMed] [Google Scholar]
- 44.Keating MJ, Flinn I, Jain V, Binet JL, Hillmen P, Byrd J, Albitar M, Brettman L, Santabarbara P, Wacker B, et al. Therapeutic role of alemtuzumab (Campath-1H) in patients who have failed fludarabine: results of a large international study. Blood. 2002;99:3554–3561. doi: 10.1182/blood.v99.10.3554. [DOI] [PubMed] [Google Scholar]
- 45.Hu W, Ge X, You T, Xu T, Zhang J, Wu G, Peng Z, Chorev M, Aktas BH, Halperin JA, et al. Human CD59 inhibitor sensitizes rituximab-resistant lymphoma cells to complement-mediated cytolysis. Cancer research. 2011;71:2298–307. doi: 10.1158/0008-5472.CAN-10-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rutkowski MJ, Sughrue ME, Kane AJ, Mills SA, Parsa AT. Cancer and the complement cascade. Mol Cancer Res. 2010;8:1453–65. doi: 10.1158/1541-7786.MCR-10-0225. [DOI] [PubMed] [Google Scholar]
- 47.Allendorf DJ, Yan J, Ross GD, Hansen RD, Baran JT, Subbarao K, Wang L, Haribabu B. C5a-mediated leukotriene B4-amplified neutrophil chemotaxis is essential in tumor immunotherapy facilitated by anti-tumor monoclonal antibody and beta-glucan. J Immunol. 2005;174:7050–6. doi: 10.4049/jimmunol.174.11.7050. [DOI] [PubMed] [Google Scholar]
- 48.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, Döhner K, Bentz M, Lichter P. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]