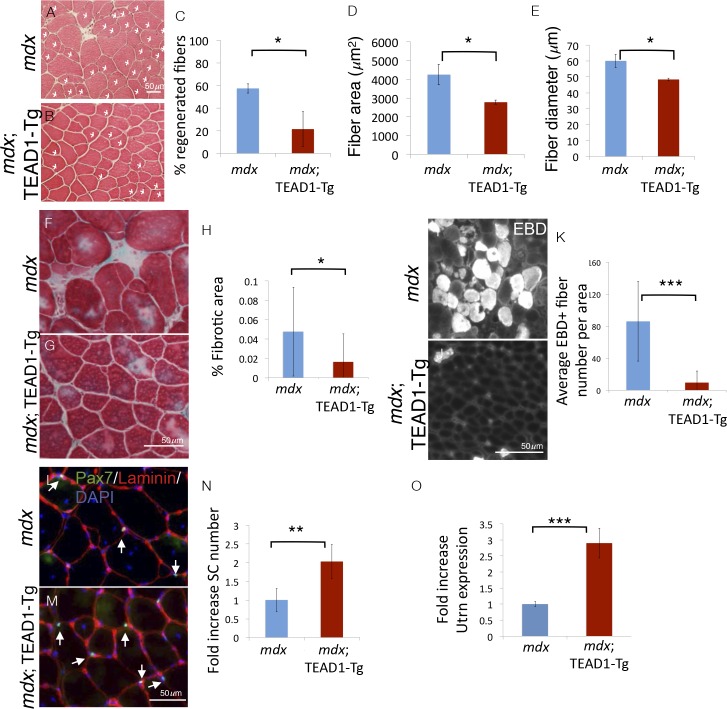
Figure 8. Pathologies of muscular dystrophy are ameliorated by TEAD1-Tg expression.
Histology, IF analyses, and qRTPCR on dystrophic muscle from mdx mice modeling chronic injury. A–C) Asterisks in H and E stained muscle sections indicate regenerated fibers in mdx (A) and mdx; TEAD1-Tg mice (B). The percent of fibers that are regenerative is quantified for each genotype in C. D–E) The fiber area (D) and fiber diameter (E) for mdx or mdx; TEAD1-Tg TA muscle is also quantified. F–H) Trichrome staining was employed to label fibrotic areas in mdx (F) or mdx; TEAD1-Tg (G) TA muscle. Percent fibrotic area was quantified (H). I–K) Membrane leakage was assessed by Evan’s Blue Dye incorporation into the myofibers of mdx (I) or mdx; TEAD1-Tg (J) TA muscle. Positive fibers per area were quantified (K). (L–N) Numbers of Pax7 expressing cells were assessed by IF of Pax7 (green), and laminin (red), counterstained with DAPI (blue) in mdx (L) or mdx; TEAD1-Tg (M). TA muscles are quantified as fold increase relative to mdx in N. Utrophin (Utrn) expression was quantified in mdx or mdx; TEAD1-Tg TA muscle via qRTPCR (O). n > 3 adult mice for all measurements, p<0.05 represented by (*), p<0.005 represented by (**), p<0.0005 represented by (***).