Fig. 4.
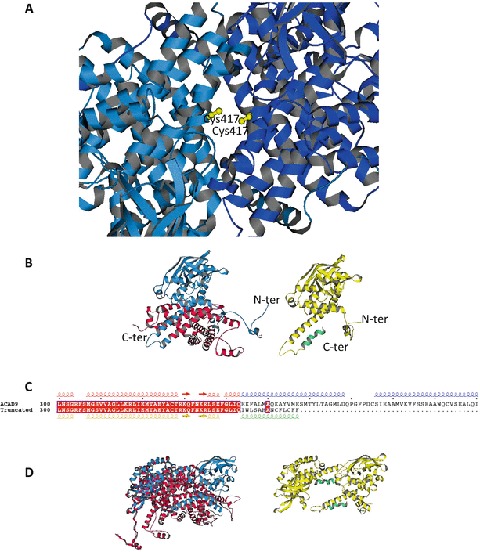
Model of the ACAD9 with the mutations c.1249C>T (p.Arg417Cys) and c.1030-1G>T. (a) ACAD9 dimer with the double c.1249C>T (p.Arg417Cys) mutation (in yellow). The ACAD9 dimer model of Nouws et al. (2010) has been used as a template. (b) Left: monomer of ACAD9 showing the truncated region (in red) resulting from the c.1030-1G>T mutation. The monomer unit is derived from the ACAD9 dimer model of Nouws et al. (2010). Right: monomer of ACAD9 showing the mutated region (in green) resulting from the c.1030-1G>T mutation. The monomer was built using the PHYRE2 server (Kelley and Sternberg 2009). (b) Sequence alignment of the 300–400 range of ACAD9 native and truncated. Secondary structural elements extracted from the models shown in a, using the same colors. (d) Left: dimer model of ACAD9 (Nouws et al. 2010) showing the lost interaction surface (in red) formed by the truncated region resulting from the c.1030-1G>T mutation. Right: dimer model of ACAD9 with the truncated and mutated monomers resulting from the c.1030-1G>T mutation. The monomers are positioned as in the native dimer
