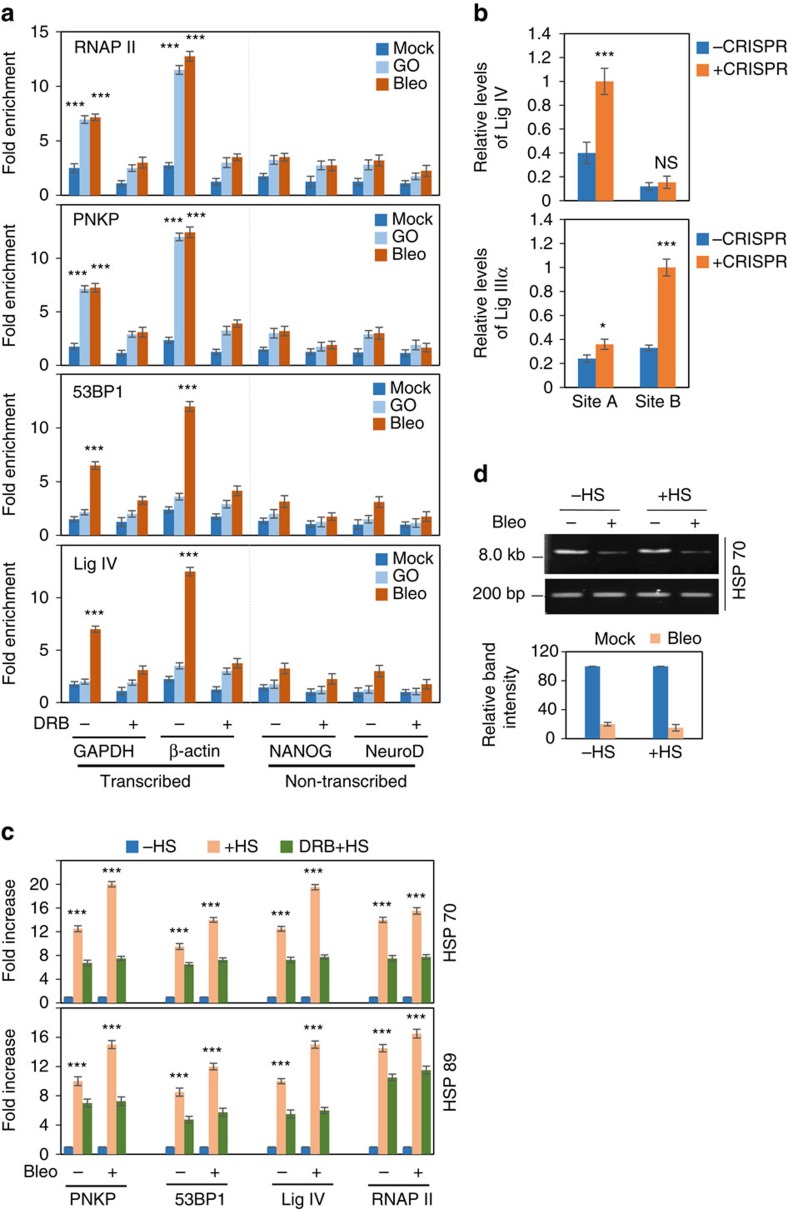
Figure 2. Quantitative ChIP assay to determine the association of C-NHEJ proteins with transcribed versus non-transcribed genes.
(a) HEK293 cells were mock-, GO- or Bleo-treated after mock (−) or DRB (+) treatment. ChIP was performed with specific Abs as indicated, and binding to the exonic regions of transcribed (GAPDH and ß-Actin) versus non-transcribed (NANOG and NeuroD) genes was quantified by qPCR from IP'd DNA. The data are represented as fold enrichment of per cent input over IgG. Error bars represent±s.d. of the mean (n≥3). ***P<0.005 represents statistical significance within a treatment group between a particular transcribed gene and both non-transcribed genes (NANOG and NeuroD) (b) DNA DSBs were generated at sites A (transcribing) and B (non-transcribing) on chromosome number 1 of human U2OS cells and ChIP was performed with Lig IV/Lig IIIα before and after CRISPR/Cas9-induction of DSBs. The data represent the relative levels of association of Lig IIIα/IV (mean±s.d.) on the DSB sites. n=3,*P<0.05; **P<0.01; ***P<0.005. (c) HEK293 cells were either mock-treated (-HS) or subjected to heat shock (+HS), or DRB treatment followed by HS (DRB+HS). The cells were further mock/Bleo-treated and ChIP was performed with specific Abs or control IgG. Binding to the HSP70 and HSP89 gene(s) was quantified by qPCR from IP'd DNA. Per cent input over IgG was calculated and represented as fold increase with -HS samples as unity. Error bars represent±s.d. of the mean (n≥3). The data are significant at ***P<0.005 between each set of –HS/+HS and +HS/DRB+HS. (d) Long amplicon quantitative PCR-mediated estimation of DNA damage in the HSP70 gene before (-HS) and after heat-shock (+HS) treatment. (–) and (+) represent mock and Bleo treatment, respectively. In each case, the short genomic fragment (∼200 bp) is amplified (SA-PCR) for normalization of the LA-qPCR data; the bar diagram represents normalized band intensity (mean±s.d.; n=3).