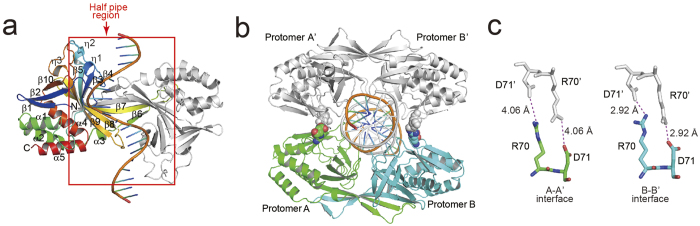
Figure 1. Overall structure of the R.PabI-nonspecific dsDNA complex.
(a) Complex structure in the asymmetric unit. One R.PabI protomer is colored blue (in the N terminus) to red (in the C-terminus). The other R.PabI protomer is colored grey. The bound ssDNA is colored orange. Secondary structure assignments of protomer A are labelled on the model. The half-pipe region is indicated by a red box. (b) Tetrameric structure of the R.PabI-nonspecific dsDNA complex. R.PabI protomers A and B in one asymmetric unit are colored green and cyan, respectively. R.PabI protomers generated by a symmetry operation (protomers A’ and B’) are colored grey. The dsDNA between the two R.PabI dimers is colored orange (chain C) and grey (chain C’). R70 and D71, which form inter-dimer salt bridges, are shown by a sphere model. (c) Salt bridges between the two R.PabI dimers. Residues of protomers A and B are shown as green and cyan stick models, respectively. Residues of protomers A’ and B’ are shown as a grey stick model. Intermolecular salt bridges are shown as magenta dotted lines.