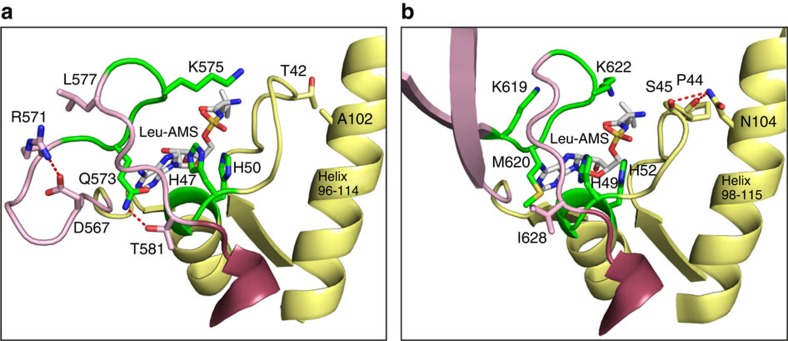
Figure 5. Interactions of key residues in the catalytic active site of AgnB2.
Ribbon structure of the catalytic active site of (a) AgnB2·tRNALeu·Leu-AMS complex and (b) E. coli·tRNALeu·Leu-AMS complex. AgnB2 key residues interacting with Leu-AMS (shown as white sticks) or adjacent to the catalytic site are depicted as sticks with the following colour code: residues of the K572QSKS576 and of the H47IGH50 catalytic motifs, are coloured in green; core permissive residues T42 and A102 are shown in yellow; T581 which H-bonds to Q573 in pink; and D567 and R571 which forms the salt bridge mutated in this study and residue L577 are also in pink. E. coli LeuRS equivalent residues in the aminoacylation complex (PDB: 4AQ7) are shown in panel b. Key interactions in both the AgnB2 and the E. coli complexes are shown as red-dashed lines.