Figure 5.
The BADC Proteins Resemble BCCP Isoforms but Are Not Biotinylated.
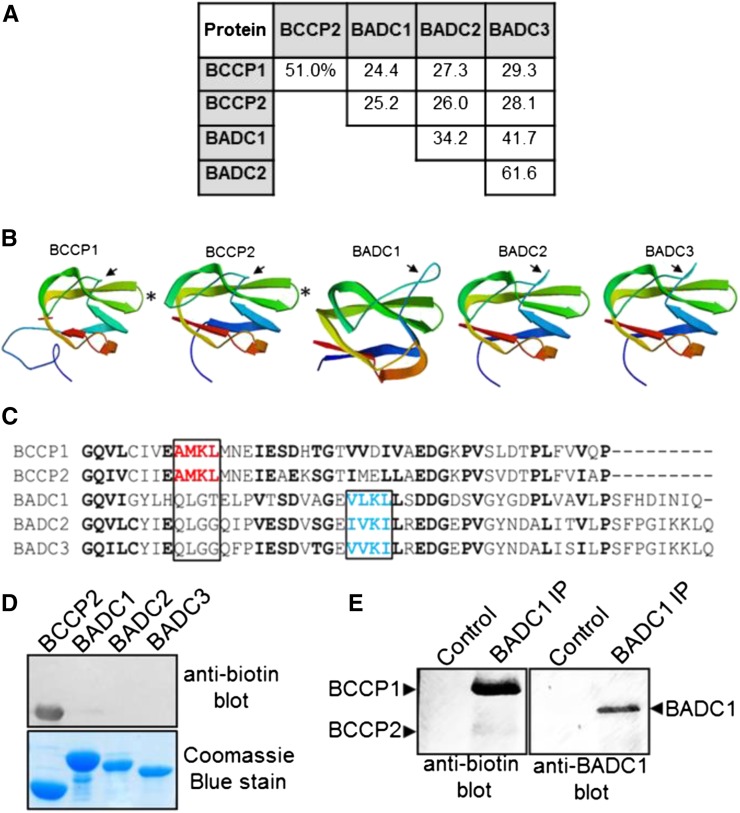
(A) Protein sequences from Arabidopsis were aligned and percent amino acid identity was calculated using ClustalW.
(B) Structures of each protein were generated using Swiss-Model. The solved structure for E. coli BCCP (PDB 4HR7) was the closest matching solved structure to all five proteins and was used as a template. Protein sequences lacking the predicted transit peptide residues were used as the input. The N and C termini are colored blue and red, respectively. Arrows indicate thumb domain. Asterisks indicate location of the biotinyl Lys residue on the BCCPs.
(C) Alignment of the C termini of the Arabidopsis BCCP and BADC proteins. Conserved residues are in bold. Highlighted in red is the canonical biotinylation motif containing the modified Lys in BCCP. Highlighted in blue is the chemically similar motif found in the BADC proteins.
(D) Protein blot analysis of recombinant Arabidopsis proteins using a biotin-specific antibody.
(E) Protein blot analysis of immunoprecipitated in vivo BADC1 from Arabidopsis seedlings. Blotting precipitate with BADC1-specific antibody showed the presence BADC1 in the sample, while blotting with biotin-specific antibody showed no recognition of BADC1.