The TCP4 transcription factor of Arabidopsis integrates developmental signals and the environmental cues to promote auxin-dependent cell elongation in hypocotyls.
Abstract
Cell expansion is an essential process in plant morphogenesis and is regulated by the coordinated action of environmental stimuli and endogenous factors, such as the phytohormones auxin and brassinosteroid. Although the biosynthetic pathways that generate these hormones and their downstream signaling mechanisms have been extensively studied, the upstream transcriptional network that modulates their levels and connects their action to cell morphogenesis is less clear. Here, we show that the miR319-regulated TCP (TEOSINTE BRANCHED1, CYCLODEA, PROLIFERATING CELL FACTORS) transcription factors, notably TCP4, directly activate YUCCA5 transcription and integrate the auxin response to a brassinosteroid-dependent molecular circuit that promotes cell elongation in Arabidopsis thaliana hypocotyls. Furthermore, TCP4 modulates the common transcriptional network downstream to auxin-brassinosteroid signaling, which is also triggered by environmental cues, such as light, to promote cell expansion. Our study links TCP function with the hormone response during cell morphogenesis and shows that developmental and environmental signals converge on a common transcriptional network to promote cell elongation.
INTRODUCTION
Cell expansion is crucial for all stages of plant development, from germination to flowering, and for growth responses to changing environmental conditions. The major plant hormones, including gibberellic acid (GA), auxin, and brassinosteroid (BR), mediate cell expansion during organ morphogenesis and environmental responses (Lau and Deng, 2010; Depuydt and Hardtke, 2011; Bai et al., 2012b; Oh et al., 2012, 2014). Auxin influences hypocotyl cell expansion by triggering the degradation of IAA3 (AUXIN/INDOLE-3-ACETIC ACID3), thereby releasing the ARF (AUXIN RESPONSE FACTOR) transcription factors, specifically ARF6 and 8, from inhibition (Vernoux et al., 2011; Oh et al., 2014). Consequently, the hypocotyl cells of the dominant shy2-2 (short hypocotyl2-2) mutant, where ARF6 is permanently sequestered by an auxin-insensitive IAA3, fail to expand even in the presence of auxin (Tian and Reed, 1999; Oh et al., 2014). BR also promotes cell expansion, among other developmental responses, by binding to the plasma membrane receptor BRI1 (BR INSENSITIVE1) and inducing downstream signal transduction, subsequently inactivating BR-INSENSITIVE2 kinase activity (Vert et al., 2005). This, in turn, results in an increased level of the de-phosphorylated BR-responsive transcription factors BZR1 (BRASSINAZOLE RESISTANT1) and BZR2/BES1 (bri1-EMS suppressor 1) (Kim and Wang, 2010), which bring about cell expansion via the upregulation of target genes, such as PACLOBUTRAZOL RESISTANT (PRE) and SMALL AUXIN UPREGULATED RNA (SAUR) (Bai et al., 2012b).
Recent studies have demonstrated that there is crosstalk between the auxin and BR signaling pathways in regulating the common target genes during cell expansion (Nemhauser et al., 2004; Oh et al., 2014). The integration of auxin and BR responses rely on ARF6-BZR1 heterodimer formation, which is indispensable for the activation of the PRE/SAUR transcriptional network that promotes cell expansion (Oh et al., 2014). A dominant form of BZR1, bzr1-1D, fails to promote cell expansion in the shy2-2 background, even in the presence of BR or auxin (Oh et al., 2014). Similarly, increased ARF6 activity by auxin-mediated degradation of IAA3 is insufficient to induce cell growth in the bri1 mutant background, where BZR1 is inactive (Nemhauser et al., 2004). Thus, both BR signaling and auxin signaling are required for the activation of the PRE/SAUR genes by BZR1-ARF heterodimer.
The role of auxin and BR in cell expansion is further supported by altered cell expansion phenotypes of mutants where the levels of these hormones are perturbed. Mutants deficient in the BR response have short stature and small cells, while BRI1 overexpression results in enlarged organs (Oh et al., 2011; Zhiponova et al., 2013). Similarly, cell expansion is perturbed in mutants with altered auxin levels. The major auxin biosynthetic pathway is dependent on endogenous tryptophan, and a key enzymatic step in this pathway is catalyzed by multiple monooxygenases encoded by the YUCCA (YUC) genes (Zhao et al., 2001; Korasick et al., 2013). The elevated auxin levels following YUC overexpression result in pleiotropic phenotypes, including increased hypocotyl length (Zhao et al., 2001).
Even though the molecular circuits involved in BR and auxin signaling and the enzymatic pathways that synthesize these hormones are known, the transcriptional network that promotes their tissue-specific expression and thereby connects their function with organ morphogenesis has started to emerge only recently (Guo et al., 2010; Sun et al., 2012; Lucero et al., 2015). It has been demonstrated that the TCP (TEOSINTE BRANCHED1, CYCLOIDEA, PROLIFERATING CELL FACTOR) family of DNA binding transcription factors plays a key role in multiple developmental processes (Martín-Trillo and Cubas, 2010). Five closely related, class II TCP genes, i.e., TCP2, 3, 4, 10, and 24, redundantly promote cell maturation during leaf morphogenesis and their transcripts are degraded by miR319 (Palatnik et al., 2003; Efroni et al., 2008). Loss of function of these genes results in enlarged leaves, due to an excess of cells that are smaller in size, while their gain of function leads to smaller leaves (Efroni et al., 2008; Sarvepalli and Nath, 2011b). However, because leaf size is regulated by a feedback mechanism in which an alteration in cell proliferation is compensated for by a corresponding but opposite effect on cell size (Hisanaga et al., 2015), the exact role of class II TCP genes on cell morphogenesis is not fully apparent. For example, it is still not clear whether the TCP genes directly promote cell expansion during organ morphogenesis.
While the transcription factors that activate auxin synthesis during environmental regulation of cell elongation have been identified (Sun et al., 2012), the proteins that promote developmentally regulated auxin synthesis during tissue morphogenesis are not known. By altering the levels of miR319-targeted TCP genes in mutant and transgenic lines, we show here that TCP4 directly activates YUC5 transcription and that this in turn integrates organ morphogenesis with auxin and brassinosteroid responses in promoting hypocotyl cell elongation.
RESULTS
TCP4 Promotes Hypocotyl Cell Elongation
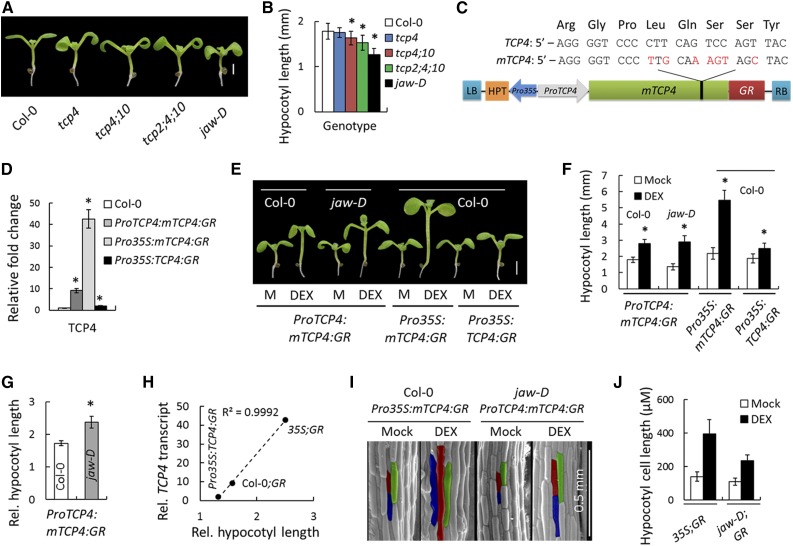
Hypocotyl growth depends on cell elongation in response to both endogenous and environmental factors and represents an ideal model to study the integration of hormone responses on cell size (Gendreau et al., 1997; Oh et al., 2014). Earlier studies from our laboratory suggested that gain of function of TCP4 enhances cell elongation (Sarvepalli and Nath, 2011b) in Arabidopsis thaliana. To examine whether TCP4 and its homologs are required for cell growth, we compared hypocotyl length in several single and multiple tcp mutants and observed that their loss of function redundantly reduced hypocotyl elongation (Figures 1A and 1B). The highest reduction was observed in the jaw-D mutant, where TCP2, 3, 4, 10, and 24 transcripts are reduced due to miR319 overexpression (Palatnik et al., 2003). To examine whether these TCP genes are sufficient for hypocotyl elongation, we generated transgenic lines in the Col-0 background in which a miR319-resistant form of TCP4 (Palatnik et al., 2003) fused to rat glucocorticoid receptor (GR) (Simon et al., 1996) was expressed under the TCP4 promoter (ProTCP4:mTCP4:GR) or 35S promoter (Pro35S:mTCP4:GR) (Figure 1C). Upon induction of TCP4 activity by external application of the GR-analog dexamethasone (DEX), the ProTCP4:mTCP4:GR seedlings showed increased hypocotyl length (Figures 1E and 1F) and reduced rosette size (Supplemental Figure 1A), compared with noninduced seedlings of the same age, which is consistent with previous results of TCP4 gain-of-function phenotypes (Schommer et al., 2008; Sarvepalli and Nath, 2011b).
Figure 1.
The miR319-Targeted TCP Genes Promote Hypocotyl Elongation.
(A) and (B) Seven-day-old seedlings (A) and their average hypocotyl lengths (B). n = 10 to 12.
(C) Schematic diagram of mTCP4:GR construct. Synonymous mutations (red) were introduced in the miR319 target region on TCP4 ORF to generate a miR319-resistant version of TCP4 (mTCP4) without changing the protein sequence (Palatnik et al., 2003). LB and RB refer to left border and right border of T-DNA, respectively; HPT, hygromycin phosphotransferase; GR, glucocorticoid receptor.
(D) Relative transcript levels of TCP4 determined by RT-qPCR analysis of 9-d-old seedlings. Average of three independent biological samples is shown. Transcript levels were normalized to that of PP2A.
(E) and (F) Seven-day-old seedlings grown in the absence (M, Mock) or presence of 12 µM DEX (E) and their average hypocotyl length (F). n = 10 to 12.
(G) Relative hypocotyl length of DEX-induced seedlings (a ratio of length under inductive condition to that under non-inductive condition). n = 12.
(H) A correlation analysis between TCP4 transcript level and relative hypocotyl length in three TCP4 gain-of-function lines.
(I) and (J) Scanning electron microscopy images of the hypocotyls of 7-d-old seedlings (I) grown in the presence or absence of 12 µM DEX and their average epidermal cell length (J). The average cell length was based on 120 to 150 cells from four hypocotyls. Representative cells are highlighted in red, green, and blue (I).
Bars = 1 mm in (A) and 2 mm in (E). Error bars indicate sd in (B), (D), (F), and (G) and se in (J). Asterisk indicates P < 0.05; unpaired Student’s t test was used to determine significant differences relative to the Col-0 values ([B], [D], and [G]) or the mock values (F). In (H) and (J), 35S;GR, Col-0;GR, and jaw-D;GR denote Pro35S:mTCP4:GR, ProTCP4:mTCP4:GR, and jaw-D;ProTCP4:mTCP4:GR plants, respectively.
To evaluate the extent to which TCP4 contributes to hypocotyl elongation, we expressed the ProTCP4:mTCP4:GR cassette in the jaw-D line, in which expression of TCP4 and its redundant homologs is markedly reduced (Palatnik et al., 2003). The resulting jaw-D;ProTCP4:mTCP4:GR transgenic line completely rescued the jaw-D phenotype upon TCP4 induction with DEX (Supplemental Figures 1B to 1E), demonstrating that TCP4 alone compensates for the loss of all miR319-targeted TCPs. However, the ProTCP4:mTCP4:GR and jaw-D;ProTCP4:mTCP4:GR plants under noninductive conditions were indistinguishable from Col-0 and jaw-D, respectively (Supplemental Figures 1A and 1B), showing that the transgenic TCP4 protein is functional only upon DEX induction. The jaw-D;ProTCP4:mTCP4:GR seedlings showed a larger relative increase in hypocotyl length upon TCP4 induction than did the ProTCP4:mTCP4:GR seedlings (Figures 1E to 1G). The increase was further enhanced upon DEX induction in the Pro35S:mTCP4:GR seedlings (Figures 1E and 1F), which expressed higher levels of TCP4 transcript (Figure 1D). By contrast, the increase in hypocotyl length was marginal in the Pro35S:TCP4:GR seedlings (Figures 1E and 1F), which expressed a miR319-susceptible form of TCP4 with lower TCP4 transcript level (Figure 1D), implying that the hypocotyl elongation is a result of miR319 activity on TCP transcripts (Figure 1H). The effect of TCP4 induction on hypocotyl length appears to be light dependent, as the dark-grown jaw-D;ProTCP4:mTCP4:GR seedlings failed to show increased hypocotyl elongation upon TCP4 induction (Supplemental Figure 2). To examine the cellular basis of hypocotyl elongation, we compared the length of hypocotyl epidermal cells in jaw-D;ProTCP4:mTCP4:GR and Pro35S:mTCP4:GR seedlings. Upon TCP4 induction, the cell length increased in both of these lines (Figures 1I and 1J). Taken together, these results provide genetic evidence that TCP4 promotes hypocotyl cell elongation, the extent to which is directly proportional to its transcript abundance (Figure 1H).
TCP4 Induction Alters the Auxin and BR Response
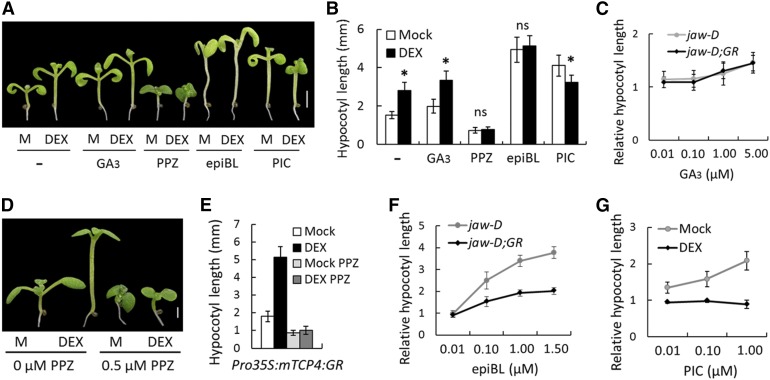
The inductive role of auxin, BR, and GA in hypocotyl elongation has been established (Oh et al., 2014). Furthermore, it has been demonstrated that the TCP genes alter the level or response of multiple plant hormones, including auxin and GA (Schommer et al., 2008; Koyama et al., 2010; Yanai et al., 2011; Das Gupta et al., 2014). To test whether the TCP4-dependent hypocotyl elongation shown above is mediated by these hormones, we monitored the effect of TCP4 induction on jaw-D;ProTCP4:mTCP4:GR hypocotyl length in the presence of these hormones or inhibitors of their responses. Even though GA3 application caused increased hypocotyl length (Figures 2A and 2B), the extent of increase was identical in the absence and presence of TCP4 induction at various GA3 concentrations (Figure 2C; Supplemental Figure 3). By contrast, BR and auxin analogs altered the effect of TCP4 induction (Figures 2A, 2B, 2F, and 2G). The BR biosynthetic inhibitor propiconazole (PPZ) (Bai et al., 2012b) inhibited the hypocotyl elongation in jaw-D;ProTCP4:mTCP4:GR (Figures 2A and 2B) and Pro35S:mTCP4:GR (Figures 2D and 2E) plants in the absence of DEX, and the inhibition could not be rescued by TCP4 induction, suggesting that TCP4 requires the BR response for promoting hypocotyl growth. Conversely, the bioactive BR epibrassinolide (epiBL) increased hypocotyl length in jaw-D seedlings in a dose-dependent manner, a response that was much reduced upon TCP4 induction (Figures 2A, 2B, and 2F), suggesting that TCP4 alters BR sensitivity. The synthetic auxin analog picloram (PIC) (Savaldi-Goldstein et al., 2008) significantly increased the hypocotyl length of jaw-D;ProTCP4:mTCP4:GR (Figures 2A and 2B) and Pro35S:mTCP4:GR (Figure 2G) plants under noninductive conditions but failed to induce any increase under TCP4-inducing conditions. The inhibition of hypocotyl growth by TCP4 activity in the presence of PIC (Figures 2A, 2B, and 2G) is consistent with previous observations that an excessive auxin response inhibits hypocotyl growth (Collett et al., 2000). These results establish a link between TCP4 function and auxin and BR responses during cell expansion.
Figure 2.
Altered Auxin and BR Responses of TCP4-Induced Hypocotyl Elongation.
(A) Seven-day-old jaw-D;ProTCP4:mTCP4:GR seedlings grown without (M, Mock) or with 12 µM DEX in the presence of 2 µM GA3, 0.5 µM PPZ, 0.4 µM epiBL, or 1 µM PIC (− indicates no chemicals).
(B) Average hypocotyl lengths of the seedlings shown in (A) (n = 10 to 15).
(C) Relative hypocotyl lengths of 7-d-old seedlings grown in the presence of 12 µM DEX and various concentrations of GA3.
(D) and (E) Eight-day-old Pro35S:mTCP4:GR seedlings (D) and their hypocotyl lengths (E) grown without (M, Mock) or with 12 µM DEX in the presence of 0.5 µM PPZ.
(F) Relative hypocotyl lengths of 7-d-old seedlings grown in the presence of 12 µM DEX and various concentrations of epiBL.
(G) Relative hypocotyl lengths of 7-d-old Pro35S:mTCP4:GR seedlings grown without (Mock) or with 12 µM DEX and various concentrations of PIC.
Bars = 2 mm in (A) and 1 mm in (D). Error bars indicate sd. Asterisk indicates P < 0.05; ns indicates not significant. An unpaired Student’s t test was used to assess significance. In (C) and (F), jaw-D;GR denotes jaw-D;ProTCP4:mTCP4:GR seedlings.
TCP4 Upregulates the Expression of Genes Involved in Auxin Biosynthesis and the Auxin/BR Response
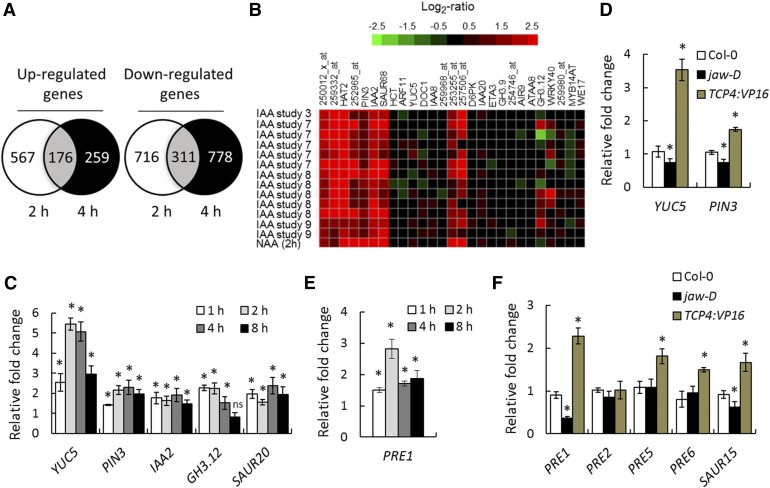
The involvement of auxin and BR in TCP4-mediated hypocotyl growth suggests that TCP4 regulates the expression of genes implicated in the biosynthesis or response of these hormones to promote cell elongation. Previous microarray studies aimed at identifying TCP targets were performed in stable mutant lines with TCP loss-of-function or in transgenic lines constitutively expressing dominant forms of TCPs (Efroni et al., 2008; Schommer et al., 2008; Sarvepalli and Nath, 2011a; Das Gupta et al., 2014). To identify the early, and therefore potentially direct, targets of TCP4 without interfering contributions from its close homologs, we compared the transcriptomes of jaw-D;ProTCP4:mTCP4:GR seedlings induced with TCP4 for 2 and 4 h with that of uninduced seedlings. The number of upregulated genes after a 2-h induction (743) was reduced (to 435) after a 4-h induction (Figure 3A), suggesting that many early targets upregulated by TCP4 return to their original level, possibly due to feedback inhibition. The number of downregulated genes remained similar after 2 h (1027) and 4 h (1089) of induction (Figure 3A).
Figure 3.
TCP4 Upregulates Auxin- and BR-Responsive Genes.
(A) Venn diagram of global transcriptome analysis of 9-d-old jaw-D;ProTCP4:mTCP4:GR seedlings treated with 12 µM DEX for 2 or 4 h.
(B) Genevestigator database (https://genevestigator.com/gv/) analysis showing that the auxin-related genes that were altered in the microarray of jaw-D;ProTCP4:mTCP4:GR plants largely overlapped with the previously reported transcriptome profiles of IAA-treated and naphthaleneacetic acid (NAA)-treated seedlings. The top 14 correlated transcriptome profiles of IAA/NAA-treated microarrays are shown. The original Genevestigator -generated figure is provided in Supplemental Figure 4.
(C) to (F) Relative transcript levels of the indicated genes determined by RT-qPCR analysis in 9-d-old jaw-D;ProTCP4:mTCP4:GR seedlings treated with 12 µM DEX for the given durations ([C] and [E]) or of the indicated genotypes ([D] and [F]). TCP4:VP16 denotes ProTCP4:TCP4:VP16 plants. All transcript levels were first normalized to PP2A/TUB2 transcripts and then the test samples were compared with the respective controls. Error bars indicate sd. Asterisk indicates P < 0.05; ns indicates not significant. An unpaired Student’s t test was used. Averages from three independent biological samples are shown.
Among the 176 upregulated and 311 downregulated genes common to the 2- and 4-h time point data (Figure 3A), 28 were related to auxin synthesis or response (Supplemental Table 1). One of these genes was YUC5, which is involved in auxin biosynthesis (Zhao et al., 2001; Cheng et al., 2006), while the remaining genes respond to external applications of auxin (Figure 3B; Supplemental Figure 4) (Nemhauser et al., 2006). RT-qPCR analysis showed that YUC5 transcript level increased ∼2.5-fold within 1 h of TCP4 induction and continued to increase until 4 h before receding (Figure 3C), suggesting that YUC5 is an early upregulated target of TCP4. Consequently, transcripts of several auxin-responsive genes, including PIN3, IAA2, GH3.12, and SAUR20, increased within 1 to 4 h of TCP4 induction (Figures 3B and 3C), suggesting that TCP4-induced YUC5 elevated the auxin level in the jaw-D;ProTCP4:mTCP4:GR seedlings. The levels of YUC5 and PIN3 were also elevated in another TCP4 gain-of-function line, ProTCP4:TCP4:VP16, in which an activated form of TCP4 is expressed under its endogenous promoter (Sarvepalli and Nath, 2011b) (Figures 3D). By contrast, YUC5 and PIN3 levels were reduced in tcp loss-of-function mutants, including jaw-D (Figure 3D; Supplemental Figure 5), implying that TCPs are required and sufficient to promote their targets involved in auxin synthesis and response.
Transcripts of several genes required for cell expansion also increased upon TCP4 induction (Supplemental Tables 1 and 2 and Supplemental Figures 6A to 6C), the prominent ones among them being PRE1 and SAUR genes. The Arabidopsis genome encodes six PRE members (PRE1 to PRE6), among which PRE1, 5, and 6 are direct targets of the BR response protein BZR1 (Oh et al., 2012). RT-qPCR validation showed that the PRE1 transcript level increased ∼3-fold within 2 h of TCP4 induction, then declined slightly and stabilized (Figure 3E). Expression of PRE1, 5, and 6 and SAUR15 (another BZR1 target) (Oh et al., 2012) increased in ProTCP4:TCP4:VP16 seedlings and that of PRE1 and SAUR15 decreased in jaw-D (Figure 3F), showing that TCP4 activates the BZR1-mediated transcriptional network responsible for cell expansion. However, transcript levels of BR biosynthetic genes (DET2) and BR-response transcription factors (BZR1, BES1, and BEH1-4) remained unaltered when TCP activity was perturbed in jaw-D or ProTCP4:TCP4:VP16 seedlings (Supplemental Figure 6B). Taken together, these results show that TCP4 promotes the expression of genes involved in auxin biosynthesis and the auxin/BR-responsive genes.
YUC5 Is a Direct Target of TCP4
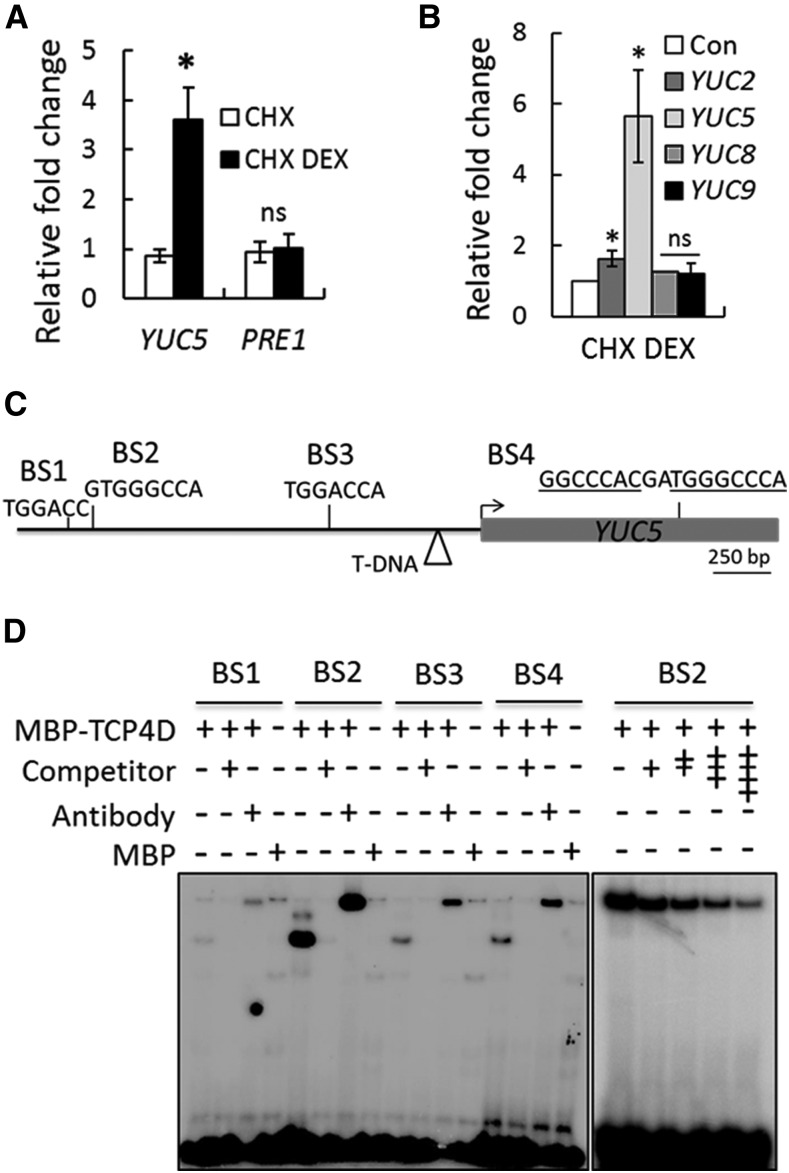
Since YUC5 expression was upregulated within an hour of TCP4 induction (Figure 3C), we examined whether YUC5 is a direct target of TCP4. Induction of TCP4 increased YUC5, but not PRE1, even in the absence of protein synthesis (Figures 4A and 4B), suggesting that TCP4 directly promotes YUC5 transcription. Sequence analysis of the YUC5 locus yielded four TCP4 binding motifs (BS1-BS4; Figure 4C) (Schommer et al., 2008). Recombinant TCP4Δ3-MBP protein, a fusion between the TCP4 DNA binding domain and maltose binding protein tag (Aggarwal et al., 2010), strongly and specifically retarded synthetic oligonucleotides corresponding to BS2, but not to other motifs in the electrophoretic mobility shift assay (Figure 4D). Induction of TCP4 resulted in increased DNA accessibility at the YUC5 locus in the chromatin context compared with the mock control (Supplemental Figure 7), as estimated by the FAIRE (formaldehyde-assisted isolation of regulatory element) experiment (Simon et al., 2012; Omidbakhshfard et al., 2014). Taken together, these results strongly suggest that TCP4 brings about changes in the chromatin of the YUC5 locus, possibly by binding to the upstream regulatory sequences directly, and promotes its transcriptional activity within hours of its induction.
Figure 4.
TCP4 Directly Upregulates YUC5.
(A) Relative transcript levels determined by RT-qPCR analysis of 9-d-old jaw-D;ProTCP4:mTCP4:GR seedlings treated either with 40 µM CHX alone or with a combination of 40 µM CHX and 20 µM DEX (CHX DEX) for 4 h.
(B) RT-qPCR analysis of the relative transcript levels in 9-d-old Pro35S:mTCP4:GR seedlings treated with 40 µM CHX or CHX plus 20 µM DEX for 4 h. Transcript levels were first normalized to PP2A ([A] and [B]) and compared with the level with only CHX treatment (Con). For both (A) and (B), error bars indicate sd. Asterisk indicates P < 0.05; ns indicates not significant. An unpaired Student’s t test was used. Averages from biological triplicates are shown.
(C) Schematic representation of the YUC5 genomic region and the four predicted TCP4 binding motifs (BS1 to BS4) in the upstream region (black line) and coding region (gray box) of the locus. Arrow indicates the predicted translation start site. The triangle indicates the T-DNA insertion site in the yuc5 line.
(D) Electrophoretic mobility shift assay gel showing retardation of radiolabeled oligonucleotides containing BS1-BS4 (shown in [C]) by recombinant TCP4Δ3-MBP protein. The + and the − symbols show the presence and the absence of the indicated compounds, respectively. A 250-fold (left panel) or 25- to 150-fold (right panel) higher concentration of unlabeled oligonucleotides was used as competitors. Anti-MBP antibody was used for the super shift reactions.
Further, the transcriptional activity of a 1.9-kb fragment corresponding to the YUC5 upstream region (Robert et al., 2013) that included the BS2 cis-element, was reduced in jaw-D;ProTCP4:mTCP4:GR X ProYUC:GUS seedlings compared with Col-0 X ProYUC5:GUS, as estimated by a GUS reporter assay under noninductive conditions (Figure 5A). However, GUS activity of the jaw-D;ProTCP4:mTCP4:GR X ProYUC5:GUS seedlings was restored to normal levels when TCP4 was induced (Figure 5A). These results, together with the data shown in Figure 3, provide biochemical evidence that TCP4 directly promotes the level of YUC5 transcript in Arabidopsis seedlings.
Figure 5.
TCP4 Induces the YUC-Dependent Auxin Response.
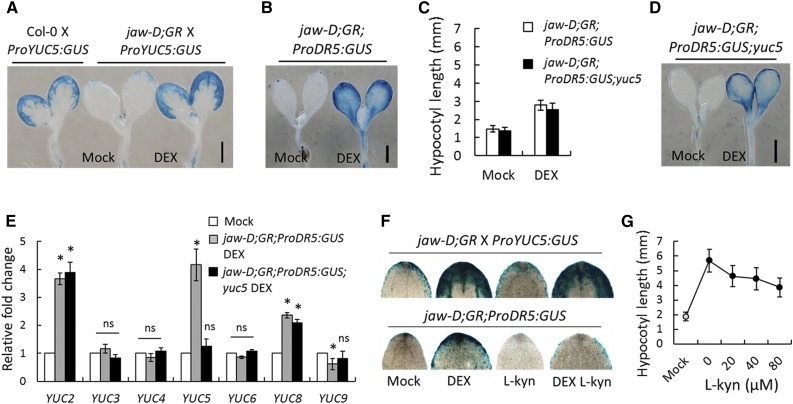
(A) and (B) GUS reporter analysis of 6-d-old seedlings grown without (Mock) or with 12 µM DEX. jaw-D;GR denotes the jaw-D;ProTCP4:mTCP4:GR genotype.
(C) Average hypocotyl length of 7-d-old seedlings grown with or without 12 µM DEX. Error bars indicate sd.
(D) GUS analysis of 6-d-old seedlings grown without (Mock) or with 12 µM DEX.
(E) Relative transcript levels determined by RT-qPCR analysis in 5-d-old seedlings (transcript abundances are provided in Supplemental Table 3). Mock and DEX indicate the absence and presence of 12 μM DEX, respectively. All transcript levels were first normalized to PP2A transcripts and then the test samples were compared with the respective controls. Error bars indicate sd. Asterisk indicates P < 0.05; ns indicates not significant. An unpaired Student’s t test was used. Averages from three independent biological samples are shown.
(F) GUS analysis of representative cotyledon tips from 6-d-old seedlings grown on medium containing the indicated chemicals; 12 µM DEX and 20 µM l-kyn were used.
(G) Hypocotyl length analysis of 8-d-old Pro35S:mTCP4:GR seedlings grown on medium containing mock (gray filled circle) or DEX (black circles) with the indicated concentrations of l-kyn. Error bars indicate sd. Bars in (A), (B), and (D) = 1 mm.
TCP4 Induces the YUC-Dependent Auxin Response in Seedlings
Increased YUC5 transcript elevates endogenous auxin levels (Woodward et al., 2005), which can be monitored by analyzing the activity of the synthetic ProDR5:GUS construct in transgenic plants (Sabatini et al., 1999). Consistent with elevated YUC5 (Figures 3 and 5A), TCP4 induction increased ProDR5:GUS activity in the jaw-D;ProTCP4:mTCP4:GR;ProDR5:GUS seedlings (Figures 5B and 5F), suggesting that TCP4 induces the auxin response in planta. To test whether the elevated auxin response is dependent on YUC5, we established a yuc5 mutant line in which the gene was disrupted by an insertion of a large T-DNA cassette 295 bp upstream of the predicted translation start site (Figure 4C; Supplemental Figure 8A). RT-PCR analysis showed that YUC5 transcript level was not abolished in yuc5 (Supplemental Figure 8B), suggesting that the T-DNA insertion did not completely disrupt the locus. Furthermore, even though TCP4 induction increased YUC5 transcript level in the jaw-D;ProTCP4:mTCP4:GR plants (Figure 3C), it failed to do so when the upstream region of YUC5 was disrupted by the TDNA insertion in the jaw-D;ProTCP4:mTCP4:GR;ProDR5:GUS;yuc5 seedlings (Figure 5E; Supplemental Table 3), suggesting that the presence of the T-DNA cassette between the BS2 element and YUC5 coding region interferes with transcriptional activation by TCP4.
The above results provide some evidence that TCP4 directly promotes YUC5 transcription. However, the yuc5 mutation failed to eliminate the TCP4-induced ProDR5:GUS activity and hypocotyl elongation in the jaw-D;ProTCP4:mTCP4:GR;ProDR5:GUS;yuc5 seedlings (Figures 5C and 5D), pointing to a YUC5-independent effect of TCP4 on modulating the auxin response, possibly via other YUC homologs. Indeed, TCP4 induction increased the level of YUC2 and YUC8 transcripts, in addition to YUC5 (Figure 5E). Moreover, previous microarray experiments showed that YUC2, 5, and 8 levels are reduced in TCP loss-of-function mutants and increased in the gain-of-function lines (Supplemental Figure 9). TCP4 induction barely increased YUC2 and failed to activate YUC8 in the absence of protein synthesis (Figure 4B), indicating that the activation of YUC2/8 by TCP4 is indirect. Recent studies demonstrated that jasmonic acid (JA) signaling upregulates YUC genes and increases endogenous auxin levels (Cai et al., 2014). It is already known that TCP4 promotes JA biosynthesis and response (Supplemental Figure 6C and Supplemental Table 1) (Schommer et al., 2008), suggesting a mechanistic basis of TCP4-induced YUC2/8 activation in jaw-D;ProTCP4:mTCP4:GR seedlings.
The action of the YUC-encoded flavin monooxygenases in the tryptophan-dependent auxin biosynthesis pathway is preceded by a key step catalyzed by tryptophan aminotransferase TAA1, which can be inhibited by the small molecule l-kynurine (l-kyn) (He et al., 2011). To assess the contribution of YUC genes to TCP4-induced hypocotyl elongation, we measured the length of DEX-induced Pro35S:mTCP4:GR hypocotyls in the presence of l-kyn. Application of l-kyn inhibited GUS activity in the jaw-D;ProTCP4:mTCP4:GR;ProDR5:GUS cotyledons in the mock condition (Figure 5F). When TCP4 was induced with DEX, addition of l-kyn markedly reduced GUS activity (Figure 5F), suggesting that the elevated auxin response upon TCP4 induction is primarily mediated by the YUC genes. The residual GUS activity in the presence of l-kyn can be explained by the YUC-independent effect of TCP4, perhaps mediated by JA (described above) (Cai et al., 2014). After downregulation of the auxin response, application of l-kyn reduced the TCP4-induced increase in hypocotyl elongation in Pro35S:mTCP4:GR seedlings in a dose-dependent manner (Figure 5G), suggesting that TCP4 is dependent on the YUC-mediated auxin response in promoting hypocotyl elongation. Taken together, these results demonstrate that TCP4 induces YUC2/5/8 transcription and the auxin response.
TCP4-Mediated Hypocotyl Growth Requires Genes Involved in the Auxin and BR Responses
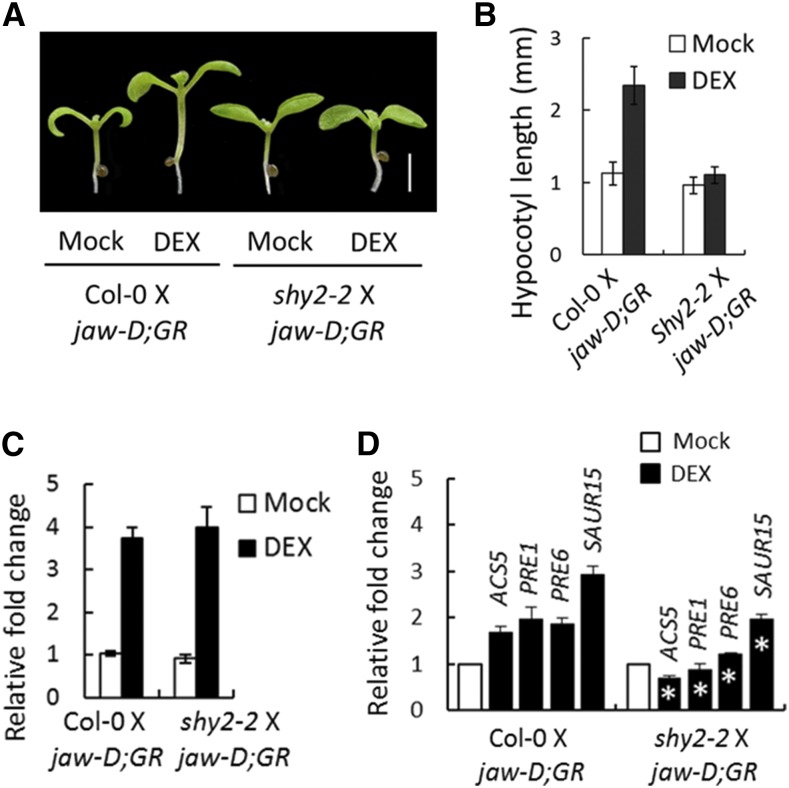
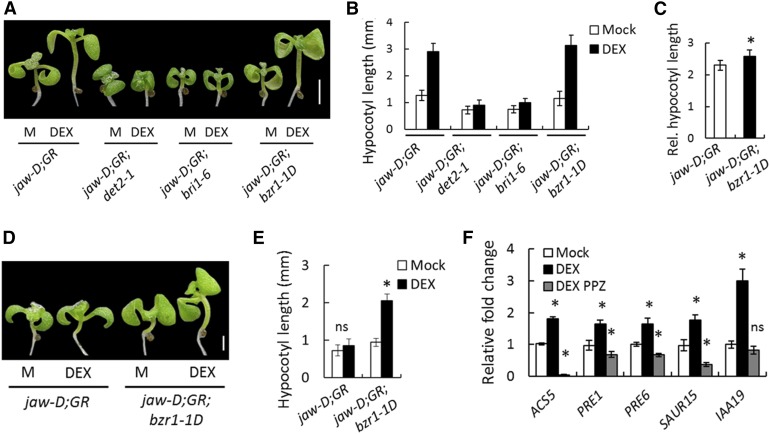
Activation of multiple YUC genes by TCP4 poses a technical challenge to genetically test whether TCP4-induced hypocotyl cell elongation is dependent on the auxin response. To overcome this, we monitored the effect of TCP4 on hypocotyl elongation by blocking the auxin response in shy2-2 seedlings, in which the auxin response is blocked due to the inactivation of IAA3/SHY2-targeted ARF proteins, notably ARF6/8 (Oh et al., 2014). The shy2-2 mutation completely suppressed the TCP4-induced elongation of jaw-D;ProTCP4:mTCP4:GR X shy2-2 hypocotyls (Figures 6A and 6B), even though higher YUC5 levels were maintained in these F1 plants (Figure 6C). These results suggest that TCP4-mediated cell elongation requires the auxin response and possibly involves auxin-induced degradation of IAA3 and the resulting release of ARF6/8 from repression. We further tested whether the activation of ARF6/8 by TCP4 requires the BR response using mutants defective in either BR synthesis (det2-1) (Noguchi et al., 1999) or signaling (bri1-6) (Li and Chory, 1997). TCP4 induction was unable to promote hypocotyl elongation when either BR biosynthesis was reduced in the jaw-D;ProTCP4:mTCP4:GR;det2-1 seedlings or BR signaling was impaired in the jaw-D;ProTCP4:mTCP4:GR;bri1-6 mutant (Figures 7A and 7B). Conversely, the dominant bzr1-1D mutation that suppresses the BR-deficient phenotype (Wang et al., 2002) significantly enhanced TCP4-induced elongation of jaw-D;ProTCP4:mTCP4:GR;bzr1-1D hypocotyls (Figures 7A to 7C). The bzr1-1D mutation also restored TCP4-mediated hypocotyl growth, even when BR synthesis was inhibited by PPZ (Figures 7D and 7E). These results demonstrate that TCP4 requires both auxin-mediated activation of ARF6/8 and BR-mediated activation of BZR1 for hypocotyl elongation, consistent with coordination between the signaling pathways of these two major hormones (Oh et al., 2014).
Figure 6.
TCP4-Induced Hypocotyl Elongation Requires Auxin Signaling.
(A) and (B) Seven-day-old seedlings grown without or with 12 µM DEX (A) and their hypocotyl lengths averaged from 10 to 12 seedlings (B). jaw-D;GR denotes the jaw-D;ProTCP4:mTCP4:GR genotype. Bar in (A) = 2 mm.
(C) Relative transcript level of YUC5 determined by RT-qPCR analysis in 10-d-old seedlings of the indicated genotypes grown without (Mock) or with 12 µM DEX.
(D) Relative transcript levels of the indicated genes determined by RT-qPCR analysis in 10-d-old seedlings of the indicated genotypes grown without (Mock) or with 12 µM DEX. Averages from three independent biological samples are shown in (C) and (D). Transcript levels were first normalized to PP2A/TUB2 and then the test samples were compared with the respective controls (Mock). Error bars indicate sd; asterisk indicates P < 0.05; an unpaired Student’s t test was used.
Figure 7.
TCP4-Induced Hypocotyl Elongation Is Dependent on BR Signaling.
(A) and (B) Seven-day-old seedlings grown without (M) or with 12 µM DEX (A) and their hypocotyl lengths averaged from 12 to 15 seedlings (B). jaw-D;GR denotes the jaw-D;ProTCP4:mTCP4:GR genotype.
(C) Relative increase in hypocotyl length upon DEX induction over mock.
(D) and (E) Seven-day-old seedlings (D) and their average hypocotyl lengths (E) of the indicated genotypes grown with or without 12 µM DEX in the presence of 250 nM PPZ. n = 10 to 12.
(F) Relative transcript level in 5-d-old jaw-D;ProTCP4:mTCP4:GR plants grown on medium supplemented with 12 µM DEX and/or 0.5 mm PPZ. Transcript levels were normalized to PP2A. n = 3.
Bars = 2 mm in (A) and 1 mm in (D). Error bars indicate sd; asterisk indicates P < 0.05. An unpaired Student’s t test was used.
The PRE and SAUR families of genes are direct downstream targets of ARF6-BZR1 heterodimer and their activation serves as a readout of ARF6-BZR1 function (Oh et al., 2014). Induction of TCP4 activated PRE1, PRE6, and SAUR15 expression, along with two other BR downstream markers, ACS5 and IAA19 (Bai et al., 2012b) (Figure 3F; Supplemental Table 2). However, their activation was significantly reduced in the DEX-induced jaw-D;ProTCP4:mTCP4:GR seedlings when BZR1 activation was inhibited by adding PPZ (Figure 7F) or in the DEX-induced jaw-D;ProTCP4:mTCP4:GR X shy2-2 seedlings (Figure 6D), in which ARF6/8 function is repressed. These results demonstrate that TCP4 requires both ARF6 and BZR1 for activating downstream cell expansion genes.
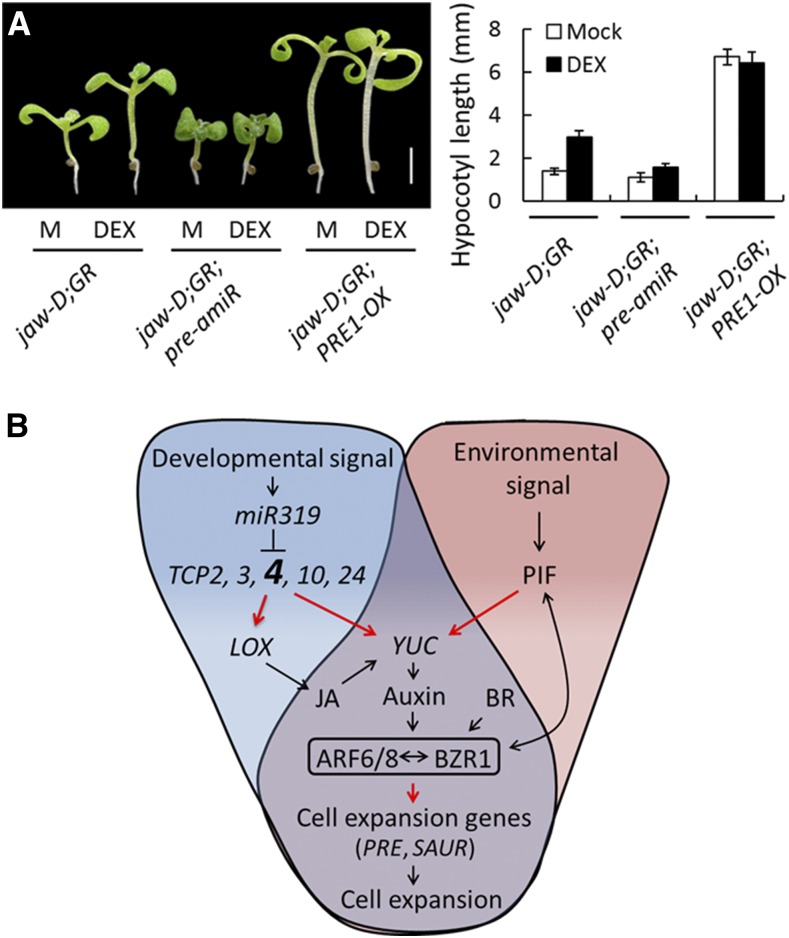
TCP4-mediated hypocotyl elongation was much reduced in the jaw-D;ProTCP4:mTCP4:GR;pre-amiR seedlings (Figures 8A and 8B), in which PRE1, 2, 5, and 6 are simultaneously downregulated due to overexpression of an artificial microRNA designed to degrade these PRE transcripts (Oh et al., 2012). On the other hand, overexpression of PRE1 markedly increased hypocotyl length in Col-0 (Bai et al., 2012a) and in jaw-D;ProTCP4:mTCP4:GR;PRE1-OX seedlings under noninductive conditions, and TCP4 induction failed to promote further hypocotyl elongation (Figures 8A and 8B), demonstrating that the PRE genes are epistatic to TCP4.
Figure 8.
TCP4 Requires PRE Genes for Hypocotyl Elongation.
(A) Seven-day-old seedlings grown without (M) or with 12 µM DEX (left) and their hypocotyl lengths averaged from 10 to 12 seedlings (right). jaw-D;GR denotes the jaw-D;ProTCP4:mTCP4:GR genotype. Bar = 2 mm. Error bars indicate sd.
(B) Schematics of a proposed pathway of miR319-targeted TCP-mediated regulation of hypocotyl cell elongation. Both the TCP-mediated developmental signal (shade of blue) and PIF-mediated environmental signal (shade of pink) converge on YUC-dependent auxin biosynthesis (shade of violet). TCP4 is enlarged and in bold, since its contribution was investigated in greater detail in this study. The resulting auxin response utilizes the common central circuit ARF-BZR1 (shown in a box) to activate the downstream regulators (such as PRE and SAUR) to promote hypocotyl cell elongation. Red arrows indicate direct transcriptional activation, black arrows indicate the direct/indirect influence of the downstream mechanism, the T-bar indicates transcript degradation, and the double-sided arrow indicates protein-protein interaction.
DISCUSSION
Cell expansion is a key event during organ morphogenesis and contributes substantially to growth and developmental plasticity in plants. It has been demonstrated that hormone responses and downstream transcription factors coordinately regulate cell size depending on developmental and environmental cues (Lau and Deng, 2010; Depuydt and Hardtke, 2011). While the molecular mechanisms mediating the environmental regulation of cell growth have been studied extensively (Bai et al., 2012a, 2012b; Oh et al., 2012, 2014), the endogenous factors that regulate this process during development are poorly understood. The TCP class of transcription factors regulates multiple developmental aspects in plants, including leaf morphogenesis, senescence, plant architecture, and circadian rhythm (Martín-Trillo and Cubas, 2010). The primary function of the class II TCP transcription factors is to promote differentiation during organ morphogenesis (Nath et al., 2003; Efroni et al., 2008; Koyama et al., 2010; Sarvepalli and Nath, 2011b). Even though several direct targets of these TCP proteins have been identified, the molecules that mediate TCP-induced cell differentiation are not known, primarily due to two major reasons. First, there is extensive functional redundancy among the class II TCP proteins, which makes their loss-of-function analysis difficult (Efroni et al., 2008; Koyama et al., 2010). Second, the gain-of-function analysis of TCP genes (Sarvepalli and Nath, 2011b) has been limited by embryonic lethality when miR319-resistant TCP transcripts are expressed even under endogenous promoters (Palatnik et al., 2003). To overcome these limitations, we expressed dexamethasone-inducible, miR319-resistant TCP4 in the jaw-D background, in which miR319-targeted TCP transcripts are much reduced.
Even though the major loss-of-function phenotypes of class II TCP genes are apparent in leaves, their gain-of-function effect on cell differentiation and maturity is seen throughout the plant (Sarvepalli and Nath, 2011b). TCP4 has been shown to repress cell proliferation to a certain extent in Arabidopsis leaves by directly promoting the cell cycle inhibitor ICK and the microRNA miR396, which degrades the transcripts of several GROWTH REGULATING FACTOR genes (Schommer et al., 2014). However, the direct effect of TCP proteins on cell expansion and maturity has not been observed in leaves. This is possibly because cell proliferation and expansion take place concomitantly in growing leaves and a loss in one is readily compensated by a gain in the other (Hisanaga et al., 2015). Therefore, the direct effect of TCP4 proteins on cell differentiation should perhaps be studied in organs in which cell expansion takes place in the absence of proliferation, such as hypocotyls (Gendreau et al., 1997). Loss of class II TCP function indeed shows reduced hypocotyl cell length, whereas their gain of function shows an increase in hypocotyl cell length (Figure 1), suggesting that TCP proteins directly promote hypocotyl cell expansion. To examine whether these TCP genes are expressed in hypocotyls, we analyzed their transcript levels in 27 independent transcriptome data sets accessed from the publicly available microarray database using Genevestigator software. The analysis indicated that these TCP genes are expressed in the hypocotyl to differing levels, with TCP2 and 24 being the most abundant (Supplemental Figure 10). However, it is possible that the TCP4-induced hypocotyl elongation is also caused by the systemic effect of increased auxin in the cotyledons and young leaf primordia (Figure 5), in addition to hypocotyl-specific TCP4.
TCP function has been associated with the biosynthesis and response pathways of several hormones in various species, including Arabidopsis. The class I TCP proteins TCP14 and TCP15 promote the cytokinin response by physically interacting with SPINDLY (Steiner et al., 2012). By contrast, the class II TCP protein TCP4 inhibits cytokinin signaling by directly promoting the expression of the type B response regulator ARR16 (Efroni et al., 2013). Furthermore, TCP4 increases the level of jasmonic acid by directly activating the LOX2 gene, which encodes an enzyme required for the rate limiting step in JA biosynthesis (Schommer et al., 2008). The class II TCP protein TCP1 of Arabidopsis increases the BR level by directly activating DWARF4, which encodes a key enzyme in BR biosynthesis (Guo et al., 2010).
Here, we demonstrate that TCP4, possibly along with other class II TCP homologs, directly promotes the transcription of the auxin biosynthesis gene YUC5 (Figure 4) and induces the auxin response in planta (Figure 5). Overexpression of YUC genes promotes the endogenous auxin level, but their single loss-of-function mutants do not show any phenotypic difference and do not alter the endogenous auxin level due to functional redundancy (Cheng et al., 2006, 2007). Based on the analysis of ProDR5:GUS activity and expression of several auxin-responsive genes (Figures 3B, 3C, and 5B), we propose that TCP4 induction increases the endogenous auxin level, which then releases the IAA3-mediated repression of ARF6/8 transcription factors to promote hypocotyl cell elongation (Vernoux et al., 2011; Oh et al., 2014). Plants with loss of function of ARF6/8 or gain of function of IAA3 show decreased hypocotyl elongation and are insensitive to auxin and BR signaling (Tian and Reed, 1999; Oh et al., 2014). We show here that TCP4 induction is also unable to promote hypocotyl growth in the absence of an auxin response in the shy2-2 lines or in the presence of L-kyn (Figures 5G and 6), an inhibitor of YUC-mediated auxin biosynthesis (He et al., 2011), suggesting that TCP4 promotes hypocotyl cell growth by promoting YUC expression and the auxin response. It was previously demonstrated that TCP4 is involved in reducing the transcription of ARF6/8 by upregulating miR167 transcription in the context of floral organ maturation (Rubio-Somoza and Weigel, 2013). Together, these findings suggest that the regulation of ARF6/8 at multiple levels constitutes an important aspect of the TCP4-mediated regulation of organ growth.
It is interesting to note that TCP3 transcriptionally activates the expression of IAA3 and negatively regulates the auxin response in planta (Koyama et al., 2010; Li and Zachgo, 2013). Conversely, TCP4 promotes the auxin-induced degradation of IAA3 protein, indicating that the miR319-targeted TCP genes maintain auxin homeostasis in different developmental aspects of plant growth. In contrast to TCP4, overexpression of the class I TCP protein TCP15 leads to a downregulation of YUC1/4 transcripts and the auxin response (Lucero et al., 2015), consistent with a proposed opposite effect of the class I and class II TCP proteins on organ growth (Li et al., 2005; Martín-Trillo and Cubas, 2010).
Interestingly, JA also promotes auxin biosynthesis by inducing ETHYLENE RESPONSE FACTOR 109, which in turn directly promotes the transcription of the auxin biosynthetic genes ANTHRANILATE SYNTHASE ALPHA SUBUNIT1 and YUC2, thereby inducing the accumulation of bioactive auxin in planta (Cai et al., 2014). JA also induces YUC8/9 expression by unknown mechanisms (Hentrich et al., 2013). The YUC proteins redundantly catalyze a rate-limiting step in the tryptophan-dependent auxin biosynthetic process (Zhao et al., 2001; Cheng et al., 2006; Mashiguchi et al., 2011; Mano and Nemoto, 2012). Besides YUC, several other genes, including AUX1/2, IAAM, and CYP79B2, also promote auxin production in a tryptophan-dependent and YUC-independent manner (Mashiguchi et al., 2011; Mano and Nemoto, 2012). Recent reports have also identified a tryptophan-independent mechanism for auxin biosynthesis (Wang et al., 2015). We observed that TCP4-induced ProDR5:GUS expression was not completely abolished in the presence of l-kyn (Figure 5F), suggesting that TCP4 promotes auxin biosynthesis in a YUC-independent manner as well, perhaps by inducing JA biosynthesis (Figure 8C).
Mutants deficient in BR and GA share common phenotypic defects, such as dwarf stature, smaller leaves with dark green color, and smaller hypocotyls and petioles (Clouse et al., 1996; Choe et al., 2001; Li et al., 2001; Oh et al., 2011). GA induces proteasome-mediated degradation of the GRAS domain-containing GA response repressors, DELLA proteins, and releases many transcription factors, including ARFs and BZR1, from their repressed state (Sun, 2011; Bai et al., 2012b; Oh et al., 2014). Even though TCP-mediated hypocotyl growth did not show an observable response to GA in our growth conditions, TCP4 triggered the activation of the ARF-BZR1 module, which is the major downstream target in GA-induced hypocotyl elongation (Bai et al., 2012b; Oh et al., 2014). Hence, we cannot ignore the possibility that GA has an indirect role in TCP-mediated hypocotyl elongation. Earlier reports have connected TCP function to GA in various plant species including Arabidopsis (Yanai et al., 2011; Steiner et al., 2012). It is interesting to note that PRE1 was initially identified as a suppressor of paclobutrazol (Lee et al., 2006), a GA biosynthesis inhibitor, and our molecular and genetic experiments show that the PRE genes are epistatic to TCP4 in promoting cell expansion (Figure 8).
Plants expressing the dominant form of BZR1, bzr11-1D, show rather a mild decrease in hypocotyl length and other BR-deficient phenotypes in the presence of light (Wang et al., 2002). These effects are complemented when the plants are treated with external BR or auxin (Oh et al., 2014). We observed that TCP4 induction also enhanced hypocotyl growth in bzr1-1D plants, similar to external auxin treatment (Figures 7A to 7C). Environmental cues such as light and temperature also regulate cell expansion during hypocotyl morphogenesis by triggering the PIF subclass of the canonical bHLH transcription factors (Sun et al., 2012). PIF4 directly promotes YUC8 expression and also physically interacts with ARF6 and BZR1 to induce the molecules that cause cell expansion, including PRE and SAUR genes (Figure 8C) (Oh et al., 2012, 2014). Our study shows that the TCP-mediated developmental signal converges on the environmental signal pathway at the level of auxin production and uses the common ARF-BZR1 central circuit to regulate cell elongation (Figure 8C). It remains to be seen whether light and temperature also influence the TCP pathway at the upstream level or whether TCP modulates the environmental pathway by physically interacting with its constituent member proteins, such as PIF, BZR, and ARF, as observed in TCP4-mediated regulation of leaf complexity (An et al., 2011).
METHODS
Plant Materials
All the genotypes are in the Col-0 background, except for bri1-6, which is in the En-2 background. The mutant lines tcp2-1 (SAIL 562-D05), tcp4-2 (GABI_363408), tcp10-2 (SALK_050423), det2-1 (CS6159), bri1-6 (CS399), and yuc5 (CS847308) were obtained from the ABRC (http://arabidopsis.org/). The ProDR5:GUS, shy2-2, bzr1-1D, jaw-D, ProTCP4:TCP4:VP16, PRE1-OX, pre-amiR, and ProYUC5:GUS lines were previously reported (Sabatini et al., 1999; Tian and Reed, 1999; Wang et al., 2002; Palatnik et al., 2003; Sarvepalli and Nath, 2011b; Bai et al., 2012a; Oh et al., 2012; Robert et al., 2013). The ProTCP4:mTCP4:GR, Pro35S:mTCP:GR, and Pro35S:TCP4:GR lines were established in the Col-0 background, and ProTCP4:mTCP4:GR was established in the indicated mutant backgrounds by crossing in the F3/F4 generation.
Establishment of DEX-Inducible TCP4 Transgenic Lines
The TCP4 open reading frame was amplified using the forward primer 5′-ATCATGTCTGACGACCAATTCCATCACC-3′ and the reverse primer 5′-CGGGATCCCGATGGCGAGAAATAGAGGA-3′, Pfu DNA polymerase, and Col-0 cDNA, and fused in frame to the rat glucocorticoid receptor, as described earlier (Simon et al., 1996), and the resulting TCP4:GR cassette was cloned into the pBSKS vector. To generate miR319-resistant TCP4, synonymous mutations were introduced in TCP4 by inverse PCR (Palatnik et al., 2003) using the forward primer 5′-CCCTTGCAAAGTAGCTACAGTCCCATGATCCGTGCTTGG-3′ and the reverse primer 5′-GTAGCTACTTTGCAAGGGACCCCTCTGAGAATACAGCTGTTGGC-3′. The resulting construct was named mTCP4:GR-KS. In parallel, a 2.16-kb-long genomic fragment corresponding to the TCP4 upstream region (ProTCP4) including the 5′ untranslated region was amplified from genomic DNA, cloned into pGEMT-Easy vector by TA cloning according to company protocol (Promega) using the forward primer 5′-AATTGACCCTTTTCTATCATGC-3′ and the reverse primer 5′-TGGTAGAGCATATTCGTCGAGA-3′ and then cloned into the pCAMBIA 1390 vector (ProTCP4-pCAMBIA 1390; Cambia; http://www.cambia.org). The mTCP4:GR fragment from mTCP4:GR-KS was cloned downstream of ProTCP4 in the ProTCP4-pCAMBIA 1390 construct to generate ProTCP4::mTCP4:GR in pCAMBIA 1390. To generate the Pro35S:mTCP4:GR cassette, the KpnI-XbaI fragment of the mTCP4:GR-KS construct was cloned downstream of the 35S promoter in the pHannibal vector and the Pro35S:mTCP4:GR cassette was cloned into the pART27 vector using NotI digestion to generate the Pro35S:mTCP4:GR cassette. Similarly, the TCP4:GR cassette was cloned downstream of the 35S promoter of pHannibal and then moved into the pART27 vector to generate Pro35S:TCP4:GR. The ProTCP4:mTCP4:GR, Pro35S:mTCP4:GR, and Pro35S:TCP4:GR constructs were integrated into the Col-0 genome by the Agrobacterium tumefaciens-mediated floral dip method (Clough and Bent, 1998).
Plant Growth Conditions and DEX Induction
Seeds were surface sterilized with 0.05% SDS dissolved in 70% ethanol for 10 min and washed two to three times with 100% ethanol. DEX stock (25 mM) was prepared in 100% ethanol and a final concentration of 12 µM DEX was added to Murashige and Skoog medium (0.5× MS salts [Sigma-Aldrich] supplemented with 0.8% phytagel and 1% sucrose) before pouring onto plates. Ethanol (0.04%) was added to the control plates and seeds were stratified for 3 d in darkness and then shifted to the growth room. For long-term DEX induction, 10-d-old seedlings were transplanted to DEX- or ethanol-treated soil and sprayed with DEX (12 µM) or ethanol (0.04%) on alternate days for 37 d. All the experiments were performed in long-day conditions (16 h white light [120 μmol/m2s]/8 h darkness) at 22°C, unless mentioned otherwise. Cyclohexamide (CHX) treatment was performed by transferring 9-d-old seedlings, without damaging the root, from MS agar medium to liquid MS medium (0.5% MS salts, 1% sucrose, and 1× vitamins; Sigma-Aldrich), which was supplemented with either 40 µM CHX (control) or 40 µM CHX and 20 µM DEX (test).
GUS Assay
GUS assays were performed as described earlier (Sessions et al., 1999; Karidas et al., 2015). Samples were collected in ice-cold 90% acetone, incubated at room temperature for 30 min, followed by washing with staining buffer (50 mM sodium phosphate, pH 7.0, 0.2% Triton X-100, 5 mM potassium ferrocyanide, and 5 mM potassium ferricyanide). Fresh staining buffer was added along with 2 mM X-Gluc and vacuum infiltrated for 30 min followed by incubation at 37°C. Staining buffer was replaced with 70% ethanol and samples were further cleared in choral hydrate:water:glycerol (8:2:1) solution, followed by mounting on glass slides. Samples were observed using an Olympus BX51 trinocular microscope fitted with a ProgRes C3 camera using ProgRes Capture Pro 2.6 software.
Phytohormone Sensitivity and Hypocotyl Length Analysis
The phytohormone sensitivity assay was performed as described earlier (Bai et al., 2012b; Oh et al., 2012). The indicated concentrations of GA3, epiBL, PPZ, l-kyn, and PIC were mixed with MS agar media before pouring onto plates. After the medium solidified, seeds were spotted on the plates with equal spacing with autoclaved toothpicks. Sown seeds were stratified for 3 d in darkness at 4°C and shifted to a plant growth chamber and kept vertically under long-day conditions at 22°C. Hypocotyls were photographed on the 7th or 8th day using a digital camera (Canon) and their lengths were measured using ImageJ software (rsbweb.nih.gov/ij/).
Microarray Experiments
Total RNA samples were isolated by the Trizol (Sigma-Aldrich) method from 9-d-old jaw-D;GR seedlings treated with DEX or mock for 2 or 4 h. After DNase I treatment, the quality of phenol-chloroform-purified RNA samples was assessed using an Agilent Bioanalyzer. RNA samples were labeled with single-color dye (Cy3), hybridized on a 8 × 60 K Arabidopsis Agilent microarray chip, and the intensity of the spots was measured and normalized according to company specifications (Agilent Technologies). Genes that underwent expression changes of ≥1.5-fold were considered to be up/downregulated (Supplemental Data Set 1). Differentially regulated genes were compared with existing TCP4 microarray data using the Genevestigator database (https://www.genevestigator.com/gv/) to identify common, differentially expressed genes.
RT-PCR Analysis
Total RNA was isolated from plant samples using the Trizol method and 1.5 µg of RNA was converted to cDNA using Revert Aid M-MuLV reverse transcriptase (Fermentas), according to the manufacturer’s instructions. PCR reactions (20 µL) were performed with the respective primers (Supplemental Data Set 2) and 33 ng of cDNA was used as template. PCR products were visualized on an ethidium bromide-stained 1% agarose gel after 30 to 33 cycles of amplification. The PCR program was set as follows: denaturation at 94°C for 30 s, annealing at 2°C above the predicted annealing temperatures, and extension at 72°C for 30 to 60 s depending on the product length. Ubiquitin was used as an internal loading control. Each reaction was repeated at least two times and one representative result is shown.
RT-qPCR Analysis
RT-qPCR was performed using a Syber Green RT-qPCR kit (KAPA SYBR FAST qPCR kits; Kapa Biosystems) in a 10-μL reaction volume according to the manufacturer’s protocol. Data were analyzed using ABI Prism 7900HT SDS software (Applied Biosystems) and the intensity ratio was calculated using the equation 2−ΔΔCT. Primer sequences are provided in Supplemental Data Set 2.
Electrophoretic Mobility Shift Assay
Oligonucleotides were end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (Thermo Fisher Scientific). The binding reaction was performed in a 15-μL reaction volume containing oligonucleotide probe, 1× binding buffer (20 mM HEPES-KOH, pH 7.8, 100 mM KCl, 1 mM EDTA, 0.1% BSA, 10 ng herring sperm DNA, and 10% glycerol) and ∼2 µg of crude recombinant protein (in the bacterial lysate). The binding reaction mixture was incubated for 30 min at room temperature and loaded onto an 8% native polyacrylamide gel. Electrophoresis was conducted at 4 V/cm for 1 h with 0.5× TBE buffer at room temperature. Gels were autoradiographed using phosphor image plate technology for 4 to 6 h. For the super shift experiment, anti-MBP monoclonal antibody (Sigma-Aldrich; catalog number M6295-2ML, lot number 122M4796) was additionally added to the binding reaction mixture to the final dilution of 25×. Lists of all primers used in this study are given in Supplemental Data Set 2.
FAIRE
FAIRE assays were performed as described earlier (Simon et al., 2012; Omidbakhshfard et al., 2014) with some modifications. Ten-day-old Pro35S:mTCP4:GR seedlings were treated with mock or 12 µM DEX for 3 h, and 2 g of tissue was fixed with formaldehyde and regulatory elements were isolated as described (Omidbakhshfard et al., 2014). TA3 was used as an internal control and TUB2 was used as a negative control. Lists of all primers used are given in Supplemental Data Set 2.
Statistical Analysis
Statistical significance was determined using an unpaired two-sample Student’s t test in GraphPad Prism software version V (GraphPad Software). The P values are indicated in the figure legends.
Accession Numbers
The Arabidopsis Information Resource (TAIR) accession numbers of the major genes used in this study are mirR319/JAW (AT4G23713), TCP2 (AT4G18390), TCP3 (AT1G53230), TCP4 (AT3G15030), TCP10 (AT2G31070), TCP24 (AT1G30210), LOX2 (AT3G45140), YUC5 (AT5G43890), and PRE1 (AT5G39860).
Supplemental Data
Supplemental Figure 1. DEX-induced TCP4 activity rescues jaw-D phenotypes.
Supplemental Figure 2. Effect of TCP4 induction on hypocotyl length in seedlings grown in darkness.
Supplemental Figure 3. Altered auxin and BR responses in TCP4-mediated hypocotyl elongation.
Supplemental Figure 4. Genevestigator analysis of auxin-related genes in the jaw-D;GR;ProTCP4:mTCP4:GR microarray.
Supplemental Figure 5. Genevestigator analysis of differentially expressed, auxin-related genes in the jaw-D;ProTCP4:mTCP4:GR microarray.
Supplemental Figure 6. Transcriptional analysis of cell expansion genes in the mutants of TCP genes.
Supplemental Figure 7. DEX-induced TCP4 promotes YUC5 promoter accessibility for transcription in vivo.
Supplemental Figure 8. Genotyping and characterization of yuc5.
Supplemental Figure 9. Genevestigator (https://genevestigator.com/gv/) analysis of YUC1 to YUC11 expression in the mutant lines of TCP genes.
Supplemental Figure 10. Genevestigator analysis of miR319-targetd TCP transcripts in wild-type hypocotyl.
Supplemental Table 1. List of auxin-related genes differentially regulated after 2 and 4 h of DEX treatment in the jaw-D;ProTCP4:mTCP4:GR microarray.
Supplemental Table 2. List of genes known to be involved in cell expansion and differentially expressed at 2 and 4 h of DEX induction in the jaw-D;ProTCP4:mTCP4:GR microarray.
Supplemental Table 3. Transcript abundance of YUC genes in TCP mutant lines.
Supplemental Data Set 1. List of genes up/downregulated upon TCP4 induction in the jaw-D;ProTCP4:mTCP4:GR seedlings.
Supplemental Data Set 2. List of primers and oligonucleotides used in this study.
Supplementary Material
Acknowledgments
We acknowledge Detlef Weigel (Max Planck Institute, Tübingen, Germany) for jaw-D, Zhi-Yong Wang (Carnegie Institute for Science, Stanford, CA) for the bzr1-1D, pre-amiR, and PRE1-OX lines, Ben Scheres (Wageningen University, The Netherlands) for the DR5::GUS line, Jiri Friml (Institute of Science and Technology, Austria) for the YUC5::GUS seeds, Jason W. Reed (University of North Carolina) for the shy2-2 seeds, the ABRC for T-DNA insertion lines, and Kavitha S. Rao (Indian Institute of Science, India) and Ashis Nandi (Jawaharlal Nehru University, India) for helpful suggestions. K.R.C. and P.A. were supported by fellowships from the Indian Institute of Science, Bangalore, and U.N. was supported by an IISc-DBT (Department of Biotechnology) Partnership grant, Government of India.
AUTHOR CONTRIBUTIONS
K.R.C. generated most of the genetic and biochemical reagents, designed and performed the experiments, analyzed the data, and wrote the first draft of the manuscript. P.A. established the ProTCP4:mTCP4:GR, Pro35S:mTCP4:GR, and Pro35S:TCP4:GR lines in the Col-0 background and performed some initial DEX induction studies. U.N. participated in designing the experiments, guided the first two authors, and finalized the manuscript.
Glossary
- BR
brassinosteroid
- DEX
dexamethasone
- GA
gibberellic acid
- PPZ
propiconazole
- epiBL
epibrassinolide
- PIC
picloram
- FAIRE
formaldehyde-assisted isolation of regulatory element
- JA
jasmonic acid
- CHX
cyclohexamide
References
- Aggarwal P., Das Gupta M., Joseph A.P., Chatterjee N., Srinivasan N., Nath U. (2010). Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 22: 1174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An L., Zhou Z., Yan A., Gan Y. (2011). Progress on trichome development regulated by phytohormone signaling. Plant Signal. Behav. 6: 1959–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.Y., Fan M., Oh E., Wang Z.Y. (2012a). A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24: 4917–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.Y., Shang J.X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.Y. (2012b). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14: 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X.T., Xu P., Zhao P.X., Liu R., Yu L.H., Xiang C.B. (2014). Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 5: 5833. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20: 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2007). Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S., Fujioka S., Noguchi T., Takatsuto S., Yoshida S., Feldmann K.A. (2001). Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 26: 573–582. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Clouse S.D., Langford M., McMorris T.C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111: 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett C.E., Harberd N.P., Leyser O. (2000). Hormonal interactions in the control of Arabidopsis hypocotyl elongation. Plant Physiol. 124: 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Gupta M., Aggarwal P., Nath U. (2014). CINCINNATA in Antirrhinum majus directly modulates genes involved in cytokinin and auxin signaling. New Phytol. 204: 901–912. [DOI] [PubMed] [Google Scholar]
- Depuydt S., Hardtke C.S. (2011). Hormone signalling crosstalk in plant growth regulation. Curr. Biol. 21: R365–R373. [DOI] [PubMed] [Google Scholar]
- Efroni I., Blum E., Goldshmidt A., Eshed Y. (2008). A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I., Han S.K., Kim H.J., Wu M.F., Steiner E., Birnbaum K.D., Hong J.C., Eshed Y., Wagner D. (2013). Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 24: 438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E., Traas J., Desnos T., Grandjean O., Caboche M., Höfte H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114: 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Fujioka S., Blancaflor E.B., Miao S., Gou X., Li J. (2010). TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22: 1161–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., et al. (2011). A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23: 3944–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentrich M., Bottcher C., Duchting P., Cheng Y., Zhao Y., Berkowitz O., Masle J., Medina J., Pollmann S. (2013). The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9 gene expression. Plant J. 74: 626–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisanaga T., Kawade K., Tsukaya H. (2015). Compensation: a key to clarifying the organ-level regulation of lateral organ size in plants. J. Exp. Bot. 66: 1055–1063. [DOI] [PubMed] [Google Scholar]
- Karidas P., Challa K.R., Nath U. (2015). The tarani mutation alters surface curvature in Arabidopsis leaves by perturbing the patterns of surface expansion and cell division. J. Exp. Bot. 66: 2107–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Wang Z.Y. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61: 681–704. [DOI] [PubMed] [Google Scholar]
- Korasick D.A., Enders T.A., Strader L.C. (2013). Auxin biosynthesis and storage forms. J. Exp. Bot. 64: 2541–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. (2010). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22: 3574–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2010). Plant hormone signaling lightens up: integrators of light and hormones. Curr. Opin. Plant Biol. 13: 571–577. [DOI] [PubMed] [Google Scholar]
- Lee S., Lee S., Yang K.Y., Kim Y.M., Park S.Y., Kim S.Y., Soh M.S. (2006). Overexpression of PRE1 and its homologous genes activates gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 47: 591–600. [DOI] [PubMed] [Google Scholar]
- Li C., Potuschak T., Colón-Carmona A., Gutiérrez R.A., Doerner P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. USA 102: 12978–12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938. [DOI] [PubMed] [Google Scholar]
- Li J., Nam K.H., Vafeados D., Chory J. (2001). BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127: 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zachgo S. (2013). TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 76: 901–913. [DOI] [PubMed] [Google Scholar]
- Lucero L.E., Uberti-Manassero N.G., Arce A.L., Colombatti F., Alemano S.G., Gonzalez D.H. (2015). TCP15 modulates cytokinin and auxin responses during gynoecium development in Arabidopsis. Plant J. 84: 267–282. [DOI] [PubMed] [Google Scholar]
- Mano Y., Nemoto K. (2012). The pathway of auxin biosynthesis in plants. J. Exp. Bot. 63: 2853–2872. [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M., Cubas P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15: 31–39. [DOI] [PubMed] [Google Scholar]
- Mashiguchi K., et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 18512–18517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U., Crawford B.C., Carpenter R., Coen E. (2003). Genetic control of surface curvature. Science 299: 1404–1407. [DOI] [PubMed] [Google Scholar]
- Nemhauser J.L., Mockler T.C., Chory J. (2004). Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2: E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser J.L., Hong F., Chory J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475. [DOI] [PubMed] [Google Scholar]
- Noguchi T., Fujioka S., Takatsuto S., Sakurai A., Yoshida S., Li J., Chory J. (1999). Arabidopsis det2 is defective in the conversion of (24R)-24-methylcholest-4-En-3-one to (24R)-24-methyl-5α-cholestan-3-one in brassinosteroid biosynthesis. Plant Physiol. 120: 833–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Bai M.Y., Arenhart R.A., Sun Y., Wang Z.Y. (2014). Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3: 03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M.H., Sun J., Oh D.H., Zielinski R.E., Clouse S.D., Huber S.C. (2011). Enhancing Arabidopsis leaf growth by engineering the BRASSINOSTEROID INSENSITIVE1 receptor kinase. Plant Physiol. 157: 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidbakhshfard M.A., Winck F.V., Arvidsson S., Riaño-Pachón D.M., Mueller-Roeber B. (2014). A step-by-step protocol for formaldehyde-assisted isolation of regulatory elements from Arabidopsis thaliana. J. Integr. Plant Biol. 56: 527–538. [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425: 257–263. [DOI] [PubMed] [Google Scholar]
- Robert H.S., Grones P., Stepanova A.N., Robles L.M., Lokerse A.S., Alonso J.M., Weijers D., Friml J. (2013). Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr. Biol. 23: 2506–2512. [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I., Weigel D. (2013). Coordination of flower maturation by a regulatory circuit of three microRNAs. PLoS Genet. 9: e1003374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Scheres B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472. [DOI] [PubMed] [Google Scholar]
- Sarvepalli K., Nath U. (2011a). Interaction of TCP4-mediated growth module with phytohormones. Plant Signal. Behav. 6: 1440–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvepalli K., Nath U. (2011b). Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 67: 595–607. [DOI] [PubMed] [Google Scholar]
- Savaldi-Goldstein S., Baiga T.J., Pojer F., Dabi T., Butterfield C., Parry G., Santner A., Dharmasiri N., Tao Y., Estelle M., Noel J.P., Chory J. (2008). New auxin analogs with growth-promoting effects in intact plants reveal a chemical strategy to improve hormone delivery. Proc. Natl. Acad. Sci. USA 105: 15190–15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C., Debernardi J.M., Bresso E.G., Rodriguez R.E., Palatnik J.F. (2014). Repression of cell proliferation by miR319-regulated TCP4. Mol. Plant 7: 1533–1544. [DOI] [PubMed] [Google Scholar]
- Schommer C., Palatnik J.F., Aggarwal P., Chételat A., Cubas P., Farmer E.E., Nath U., Weigel D. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A., Weigel D., Yanofsky M.F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20: 259–263. [DOI] [PubMed] [Google Scholar]
- Simon J.M., Giresi P.G., Davis I.J., Lieb J.D. (2012). Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat. Protoc. 7: 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R., Igeño M.I., Coupland G. (1996). Activation of floral meristem identity genes in Arabidopsis. Nature 384: 59–62. [DOI] [PubMed] [Google Scholar]
- Steiner E., Efroni I., Gopalraj M., Saathoff K., Tseng T.S., Kieffer M., Eshed Y., Olszewski N., Weiss D. (2012). The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24: 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Qi L., Li Y., Chu J., Li C. (2012). PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 8: e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T.P. (2011). The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 21: R338–R345. [DOI] [PubMed] [Google Scholar]
- Tian Q., Reed J.W. (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126: 711–721. [DOI] [PubMed] [Google Scholar]
- Vernoux T., et al. (2011). The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 7: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G., Nemhauser J.L., Geldner N., Hong F., Chory J. (2005). Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 21: 177–201. [DOI] [PubMed] [Google Scholar]
- Wang B., Chu J., Yu T., Xu Q., Sun X., Yuan J., Xiong G., Wang G., Wang Y., Li J. (2015). Tryptophan-independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 112: 4821–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513. [DOI] [PubMed] [Google Scholar]
- Woodward C., Bemis S.M., Hill E.J., Sawa S., Koshiba T., Torii K.U. (2005). Interaction of auxin and ERECTA in elaborating Arabidopsis inflorescence architecture revealed by the activation tagging of a new member of the YUCCA family putative flavin monooxygenases. Plant Physiol. 139: 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai O., Shani E., Russ D., Ori N. (2011). Gibberellin partly mediates LANCEOLATE activity in tomato. Plant J. 68: 571–582. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Christensen S.K., Fankhauser C., Cashman J.R., Cohen J.D., Weigel D., Chory J. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309. [DOI] [PubMed] [Google Scholar]
- Zhiponova M.K., et al. (2013). Brassinosteroid production and signaling differentially control cell division and expansion in the leaf. New Phytol. 197: 490–502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.