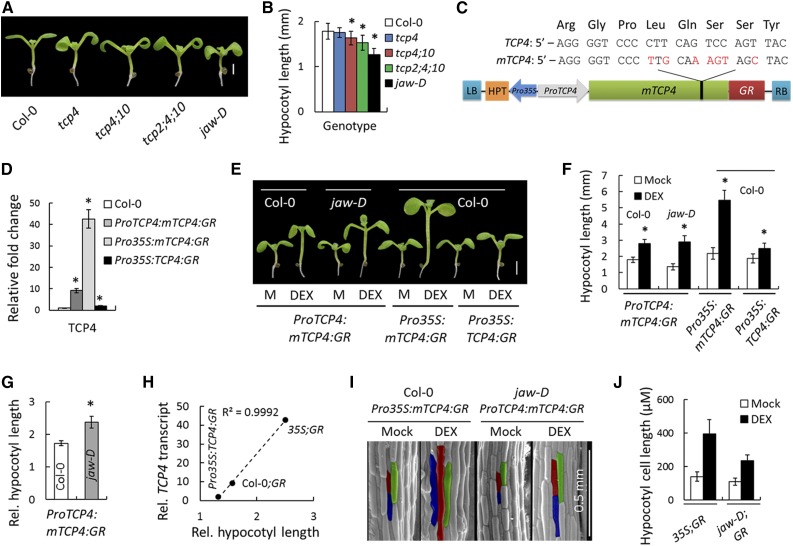
Figure 1.
The miR319-Targeted TCP Genes Promote Hypocotyl Elongation.
(A) and (B) Seven-day-old seedlings (A) and their average hypocotyl lengths (B). n = 10 to 12.
(C) Schematic diagram of mTCP4:GR construct. Synonymous mutations (red) were introduced in the miR319 target region on TCP4 ORF to generate a miR319-resistant version of TCP4 (mTCP4) without changing the protein sequence (Palatnik et al., 2003). LB and RB refer to left border and right border of T-DNA, respectively; HPT, hygromycin phosphotransferase; GR, glucocorticoid receptor.
(D) Relative transcript levels of TCP4 determined by RT-qPCR analysis of 9-d-old seedlings. Average of three independent biological samples is shown. Transcript levels were normalized to that of PP2A.
(E) and (F) Seven-day-old seedlings grown in the absence (M, Mock) or presence of 12 µM DEX (E) and their average hypocotyl length (F). n = 10 to 12.
(G) Relative hypocotyl length of DEX-induced seedlings (a ratio of length under inductive condition to that under non-inductive condition). n = 12.
(H) A correlation analysis between TCP4 transcript level and relative hypocotyl length in three TCP4 gain-of-function lines.
(I) and (J) Scanning electron microscopy images of the hypocotyls of 7-d-old seedlings (I) grown in the presence or absence of 12 µM DEX and their average epidermal cell length (J). The average cell length was based on 120 to 150 cells from four hypocotyls. Representative cells are highlighted in red, green, and blue (I).
Bars = 1 mm in (A) and 2 mm in (E). Error bars indicate sd in (B), (D), (F), and (G) and se in (J). Asterisk indicates P < 0.05; unpaired Student’s t test was used to determine significant differences relative to the Col-0 values ([B], [D], and [G]) or the mock values (F). In (H) and (J), 35S;GR, Col-0;GR, and jaw-D;GR denote Pro35S:mTCP4:GR, ProTCP4:mTCP4:GR, and jaw-D;ProTCP4:mTCP4:GR plants, respectively.