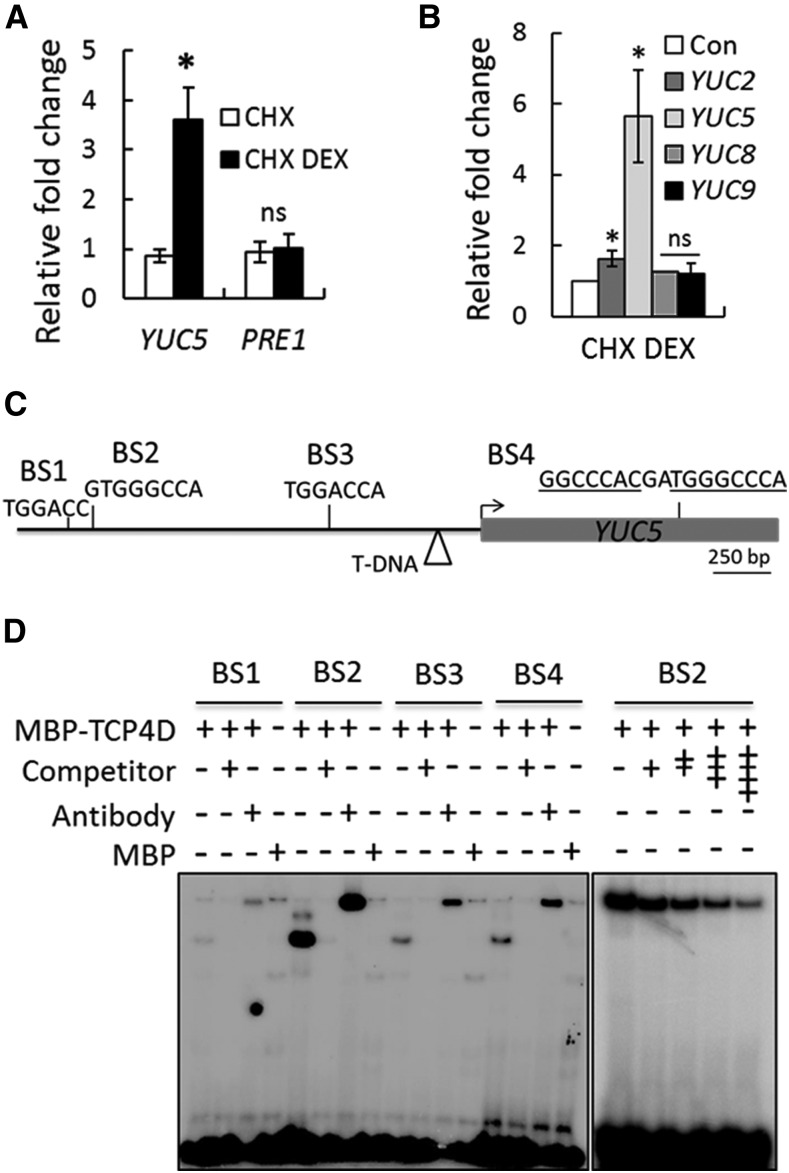
Figure 4.
TCP4 Directly Upregulates YUC5.
(A) Relative transcript levels determined by RT-qPCR analysis of 9-d-old jaw-D;ProTCP4:mTCP4:GR seedlings treated either with 40 µM CHX alone or with a combination of 40 µM CHX and 20 µM DEX (CHX DEX) for 4 h.
(B) RT-qPCR analysis of the relative transcript levels in 9-d-old Pro35S:mTCP4:GR seedlings treated with 40 µM CHX or CHX plus 20 µM DEX for 4 h. Transcript levels were first normalized to PP2A ([A] and [B]) and compared with the level with only CHX treatment (Con). For both (A) and (B), error bars indicate sd. Asterisk indicates P < 0.05; ns indicates not significant. An unpaired Student’s t test was used. Averages from biological triplicates are shown.
(C) Schematic representation of the YUC5 genomic region and the four predicted TCP4 binding motifs (BS1 to BS4) in the upstream region (black line) and coding region (gray box) of the locus. Arrow indicates the predicted translation start site. The triangle indicates the T-DNA insertion site in the yuc5 line.
(D) Electrophoretic mobility shift assay gel showing retardation of radiolabeled oligonucleotides containing BS1-BS4 (shown in [C]) by recombinant TCP4Δ3-MBP protein. The + and the − symbols show the presence and the absence of the indicated compounds, respectively. A 250-fold (left panel) or 25- to 150-fold (right panel) higher concentration of unlabeled oligonucleotides was used as competitors. Anti-MBP antibody was used for the super shift reactions.