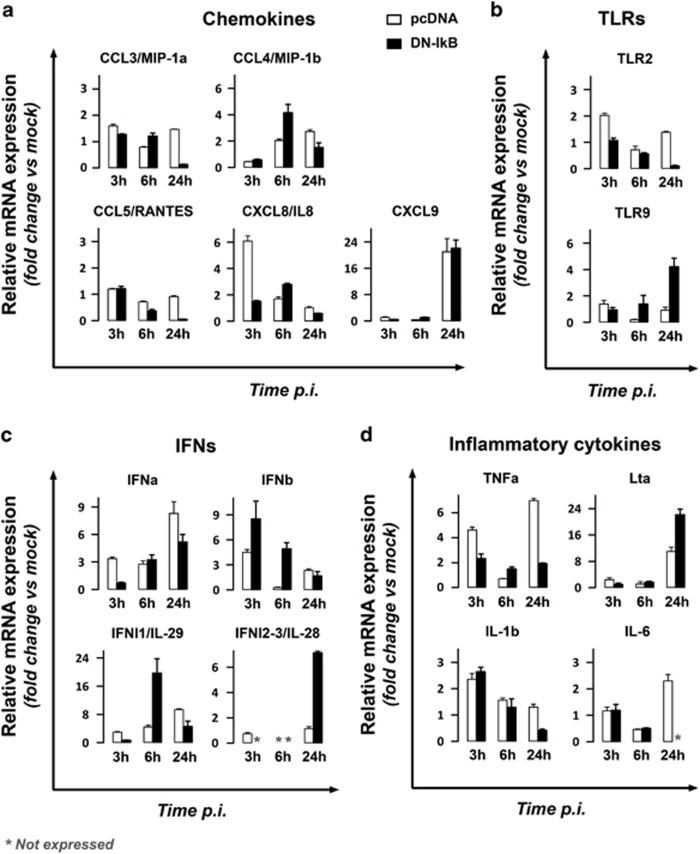
Figure 7.
Gene profiling of the antiviral innate response in HSV-1 infected U937-pDNA and U937-DN-IκB using real-time RT-PCR. Total RNA was isolated and reverse transcribed from cells infected with 50 MOI of HSV-1 for 3, 6 and 24 h and respective time-matched mock-infected control cells. Changes in transcripts were quantified by real-time RT-PCR. For each transcript of interest, data were normalized to 18 S rRNA and results were expressed as fold change in mRNA expression of infected cells compared to time-matched mock-infected cells. Bars represent mean values±s.d. from two independent experiments performed in duplicates. Statistical significances of the pcDNA versus the corresponding DN-IkB group, obtained by Bonferroni's post-hoc ANOVA, for each group of genes are the following: (a) Chemokines. CCL3: at all times assayed, P<0.001. CCL4: t3, not significant; t6, P<0.001; t24, P=0.005. CCL5: t3, not significant; t6 and t24, P<0.001. CXCL8: t3 and t6, P<0.001; t24, not significant. CXCL9: at all times assayed, not significant. (b)Toll-like-receptors (TLRs). TLR2: t3 and t24, P<0.001; t6, not significant. TLR9: t3, not significant; t6, P<0.047; t24, P<0.001. (c) Interferons (IFNs). IFN-alpha (IFNa): t3, P<0.006; t6, not significant; t24, P=0.001. IFN-beta (IFNb): t3, P<0.004; t6, P=0.001; t24, not significant. IFN-lambda1 (IFNl1): t3 and t24, not significant; t6, P<0.001. IFN-lambda2 (IFNl2): t24, P<0.001. (d) Pro-inflammatory cytokines. TNF-alpha (TNFa): t3 and t24, P<0.001; t6, P=0.007. Lymphotoxin alpha (Lta): t3 and t6, not significant; t24,P<0.001. IL-1beta (IL-1b): t3 and t6, not significant; t24,P=0.001. IL-6: t3 and t6, not significant