Fig. 2.
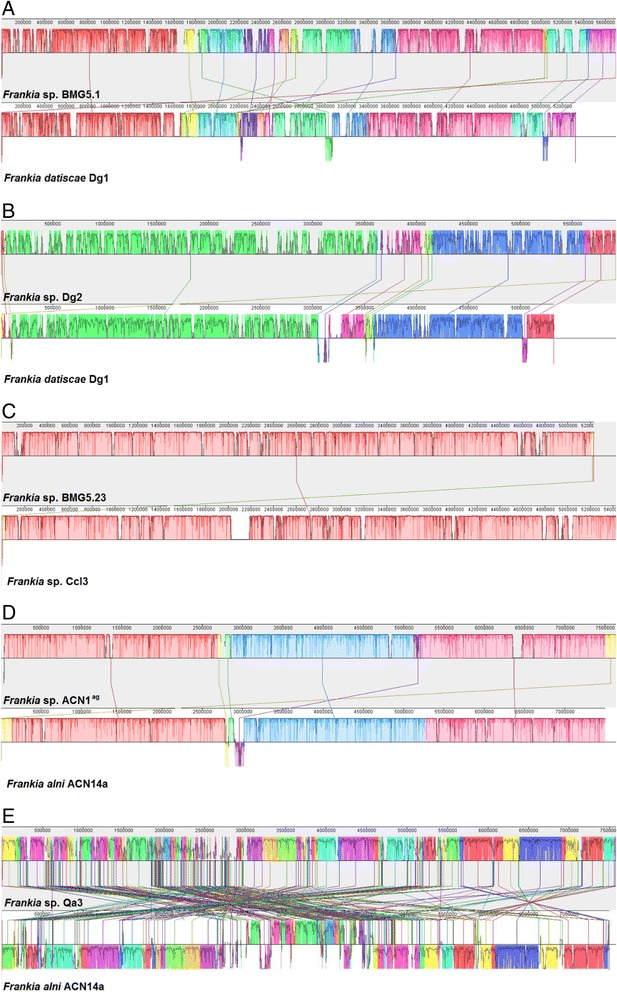
Frankia genome alignments using MAUVE [66]. Chromosome coordinates are plotted on the x-axis and the y-axis denotes the percentage of sequence identity. Colored areas appear above and possibly below the center line. Each of these areas surrounds a region of the genome sequence that aligned to part of another genome, and is presumably homologous and internally free from genomic rearrangement. When an area lies above the center line the aligned region is in the forward orientation relative to the first genome sequence. Areas below the center line indicate regions that align in the reverse complement orientation. Regions outside the colored areas lack detectable homology among the input genomes. Inside each area, Mauve draws a similarity profile of the genome sequence. The height of the similarity profile corresponds to the average level of conservation in that region of the genome sequence. Regions that are completely white were not aligned and probably contain sequence elements specific to a particular genome. The height of the similarity profile is calculated to be inversely proportional to the average alignment column entropy over a region of the alignment. a Pairwise alignment of the genomes of Candidatus Frankia datiscae Dg1 (chromosome) and Candidatus Frankia datiscae Dg2 (1066 scaffolds) (88 % ANI). b Pairwise alignment of the genomes of Candidatus Frankia datiscae Dg1 (chromosome) and Frankia sp. BMG5.1 (116 scaffolds) (96 % ANI). c Pairwise alignment of the genomes of Frankia sp. CcI3 (chromosome) and Frankia sp. BMG5.23 (166 scaffolds) (99 % ANI). d Pairwise alignment of the genomes of Frankia alni ACN14a (chromosomes) and Frankia sp. ACN1ag (90 scaffolds) (92 % ANI). e Pairwise alignment of the genomes of F. alni ACN14a (chromosome) and Frankia sp. QA3 (chromosome) (91 % ANI)
