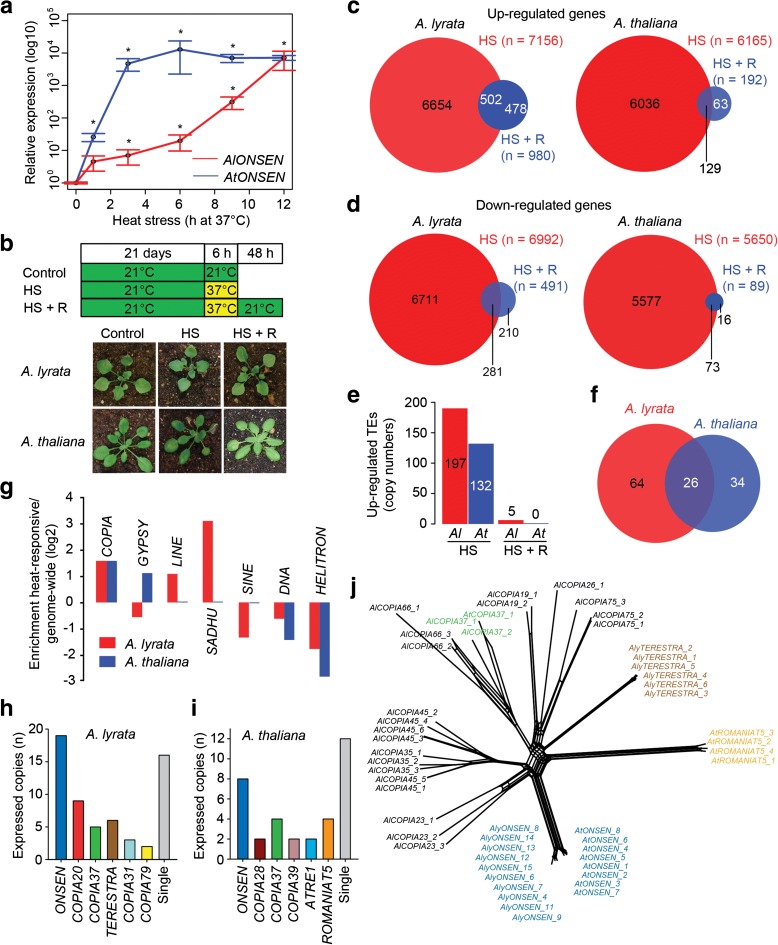
Fig. 1.
Transcriptome analysis of heat-stressed A. lyrata and A. thaliana plants. a Effects of HS on ONSEN heat-responsiveness in A. thaliana and A. lyrata. Both species were stressed at 37 °C for the indicated number of hours (h) and subsequently analyzed for the amount of ONSEN transcript (log10) by RT-qPCR relative to GAPD-H transcript amounts. * significant (t-test, P <0.05) transcript enrichment relative to 0 h control. Error bars indicate standard deviation of three biological replicates. b Design of plant HS treatment for RNA-seq and representative phenotypes of control, 6 h heat-stressed at 37 °C and recovered plants. c, d Number of significantly (c) upregulated or (d) downregulated protein-coding genes after 6 h at 37 °C and 48 h recovery at non-stress conditions in both species. e, f Number of significantly upregulated (e) TEs and (f) TE families after 6 h HS and 48 h recovery. g Identification of TE groups enriched for heat-responsive copies. Retrotransposons were divided into SINE, SADHU, LINE, COPIA, and GYPSY family members. The relative enrichment of heat-activated TEs was calculated as ratio between % of all heat-activated to % of all TEs genome-wide and expressed on a log2 scale. The major heat-responsive COPIA families in (h) A. lyrata and (i) A. thaliana. The families containing a single HRE are displayed as “single copies.” j RT amino acid sequences (Additional file 12)-based phylogenetic network of selected heat-responsive (colored) and non-responsive (black) A. lyrata and A. thaliana COPIA families. The data are also provided as un-rooted three in Additional file 5: Figure S2