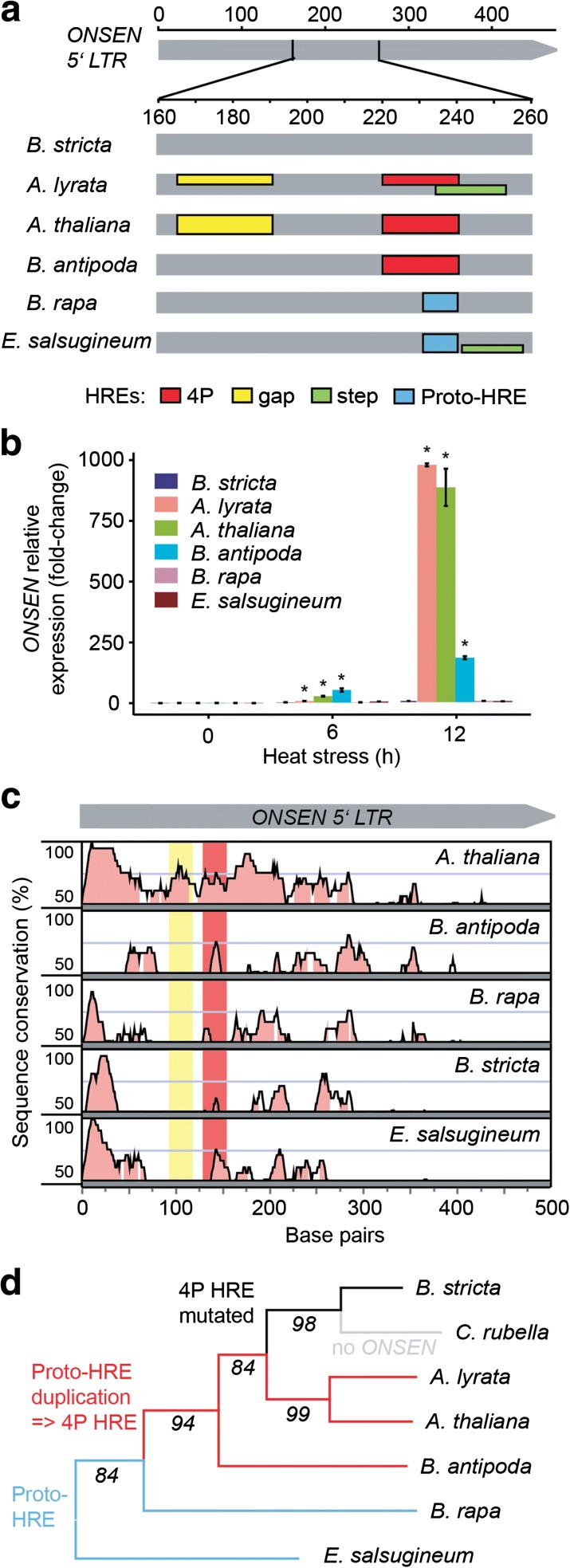
Fig. 2.
Evolution of ONSEN heat-responsiveness. a Schematic representation of in silico reconstruction of putative HREs in ONSEN 5’ LTR in different Brassicaceae species. HRE reconstruction follows criteria proposed by [20]. Colored boxes spanning the entire height of the gray field indicate HREs found in ≥50 % of the heat-responsive copies in A. thaliana and A. lyrata or all copies in other species. The lower boxes represent less frequent (<50 %) variants. Detailed information including sequences underlying individual HREs can be found in Additional file 5: Figure S2. b Transcript levels of ONSEN elements in Brassicaceae after 6 h and 12 h at 37 °C. Quantitative PCR values were obtained using species-specific primer pairs and normalized to UBC28. Error bars indicate standard deviation of three biological replicates and * P <0.05 in Student’s t-test. c Sequence conservation over the ONSEN 5’ LTR. Species-specific 5’ LTR consensus sequences were compared to A. lyrata query using 20 bp sliding window and 7 bp minimum consensus length. The y-axis for each species shows 50–100 % sequence conservation. Regions with ≥70 % similarity (pink-filled) were considered as conserved. Red and yellow background colors indicate the A. lyrata 4P and gap HRE regions. d Reconstruction of ONSEN HRE evolution. The phylogenetic tree was developed using the CHALCONE SYNTHASE gene of each individual species. The numbers at branches indicate bootstrap values. Blue lines show species with proto-HREs, red shows those carrying 4P HREs, black shows loss of HREs in B. stricta, and gray shows the COPIA78 family in C. rubella