Figure 4.
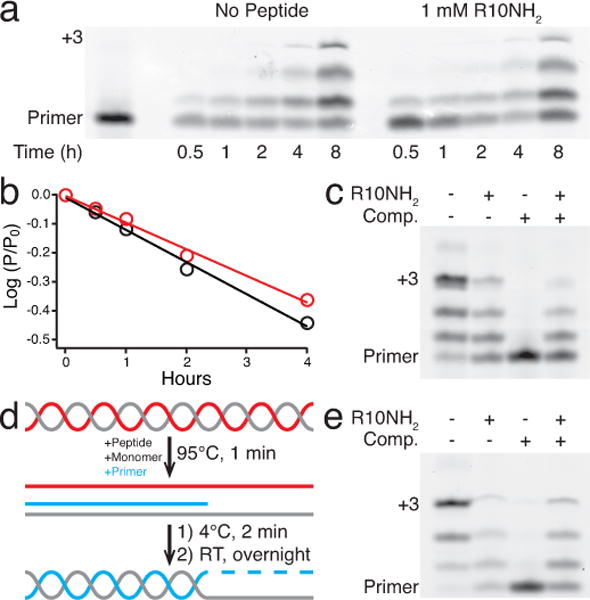
Nonenzymatic RNA polymerization. Reactions performed with 10 mM MgCl2, 250 mM Na-HEPES pH 8, and 10 mM 2-MeImpG. a) Polyacrylamide gel of nonenzymatic additions of 2-MeImpG to a 5′-cyanine 3 (Cy3)-labeled primer (5′-Cy3-CAGACUGG-3′, 2 μM) on a C4 template (5′-AACCCCCCAGUCAGUC-3′, 2.5 μM) with or without 1 mM R10NH2. Bolded cytosines represent 2-MeImpG binding sites on the template. b) log of the fraction of unreacted primer for gel lanes in a vs. time, with (red) or without (black) R10NH2. The slope of the lines (linear fit, R2 = 0.99) represents the pseudo-first-order rate constant, kobs, in h−1 for the respective reactions. With 1 mM R10NH2, kobs = 0.084(5) h−1. With no peptide, kobs = 0.110(5) h−1. SEM in parentheses. n = 5. c) Polyacrylamide gel of an overnight nonenzymatic primer extension experiment with 1.2 μM primer and 1.25 μM template (incubated with or without 1.5 mM R10NH2), after addition of 0 μM or 2 μM complementary strand to the template (5′-GACUGACUGGGGGGUU-3′) separately incubated with or without 1.5 mM R10NH2. d) Post-replication round of nonenzymatic primer extension. The template (gray) and its complement (red) were annealed, then peptide, monomer, and primer (blue) were added and the mixture was heated briefly to 95 °C. After cooling to 4 °C to allow primer-template binding, the system was allowed to warm to room temperature. e) Polyacrylamide gel of the overnight primer extension experiment described in d with 0.95 μM primer, 1 μM template, and with or without 1.2 μM complementary strand and 1.5 mM R10NH2, respectively.
