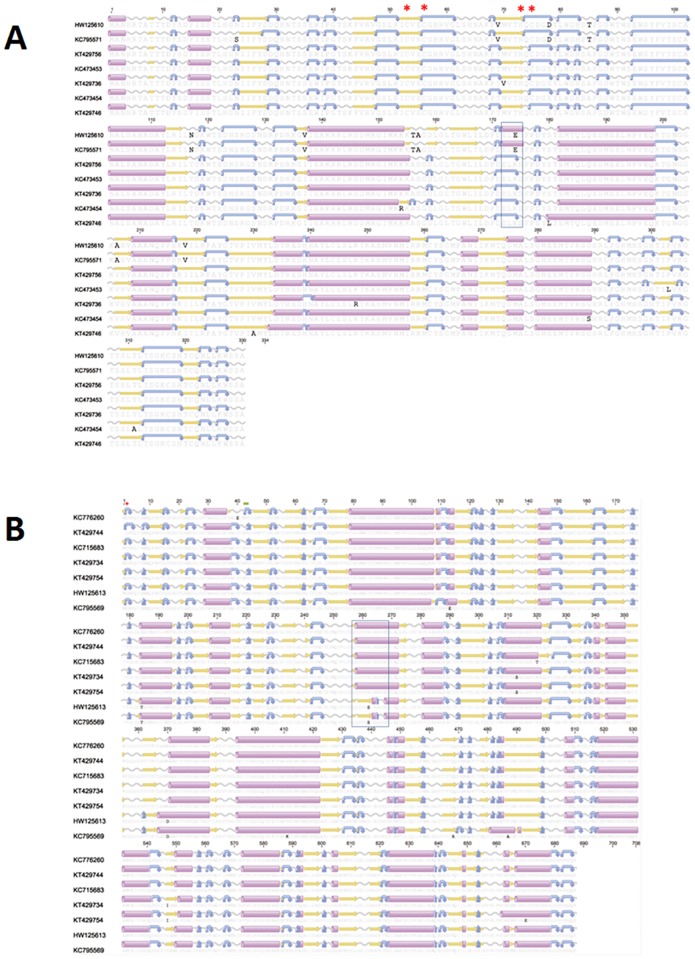
Fig 6. Amino acid alignment of open reading frame consensus sequences encoding the Piscine reovirus σ3 and μ1 protein.
Secondary structure and transmembrane domains were predicted using EMBOSS 6.6.7 (Geneious software v6.1). Predicted secondary structure of alpha helix, beta strand, coil and turn are presented in purple cylinders, yellow arrows, grey sinusoids and blue curved arrow. Sequences are identified using the GenBank accession numbers. A/ represents ORF sequences encoding PRV σ3 amino acid alignment. Red stars are conserved Zn-finger motifs. B/ represents ORF sequences encoding PRV μ1 amino acid alignment. Red cross is myristoylation site in the MRV protein and green line is post-translational cleavage site in MRV and ARV [7].