Figure 4.
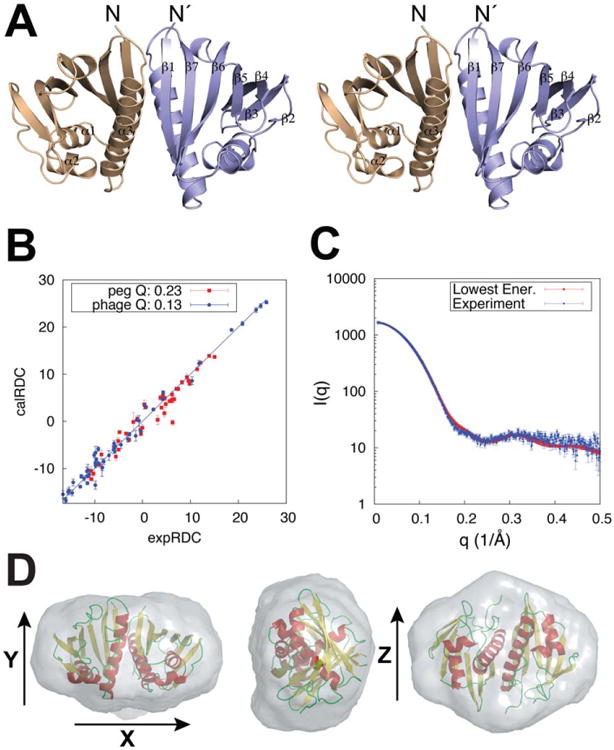
Final model fit to the RDC and SAXS data. A) Stereo view of the Aha1 symmetric dimer structure with secondary structure elements indicated for a single chain. B) Calculated versus experimental 1H-15N RDCs showing the agreement of the minimally restrained CS-Rosetta lowest energy structure to the experimental data. C) SAXS data and corresponding fit obtained for the lowest energy structure (χ2 = 1.71). D) Ab initio molecular envelope from a consensus model of 20 individual reconstructions shown along x, y, and z vectors. Spatial discrepancy of 0.540 and variation of 0.023 were obtained in the final fit. The final structural ensemble was selected using energy-based criteria (Fig. 3 and Supporting Information Fig. S8) and further validated using an independent dataset of inter-molecular NOEs [Fig. 5(B)].
