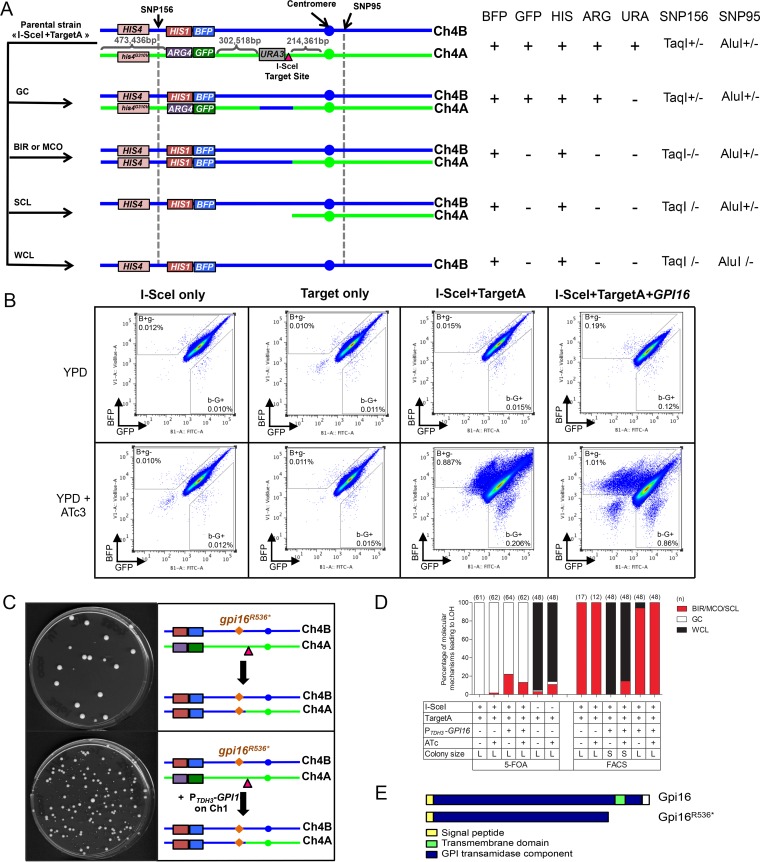
FIG 2 .
Effect of I-SceI induction in the I-SceI+TargetA and control strains. (A) Different LOH events on Chr4A can arise from I-SceI. As seen in Fig. 1A, the combined heterozygosity or homozygosity of SNP156 and SNP95 gives insights about the molecular mechanisms leading to LOH events. GC, gene conversion; BIR, break-induced replication; MCO, mitotic crossover; SCL, segmental chromosome loss; WCL, whole-chromosome loss. (B) Co-occurrence of I-SceI and its target sequence triggers a predominant increase in levels of mono-BFP cells. Data represent 106 events. The B+g− and b−G+ gates were defined arbitrarily. (C) Integration of the full-length allele of GPI16 on Chr1 allows recovery of viable mono-BFP cells after cell sorting. While cells obtained from strain I-SceI+TargetA showed poor viability due to homozygosis of the gpiR536* allele, complementation with a wild-type GPI16 allele in strain I-SceI+TargetA+GPI16 restored viability. The largest colonies observed in both cases were doubly fluorescent, having not undergone I-SceI cleavage on Chr4A. (D and E) SNP-RFLP analysis showed that I-SceI-dependent DNA DSBs on Chr4A are mainly repaired by GC. Histograms present the proportion of BIR/MCO or SCL, GC, and WCL events in the population having undergone a LOH recovered either from 5-FOA counterselection (D) or from FACS analysis (E). BIR/MCO or SCL events correspond to mono-BFP cells that displayed a homozygous SNP156 but have maintained a heterozygous SNP95. WCL events correspond to mono-BFP cells in which both SNP156 and SNP95 became homozygous. GC events correspond to doubly fluorescent cells in which both SN156 and SNP95 remained heterozygous. L, large-sized colonies; S, small-sized colonies. (F) The gpi16R536* allele might result in the truncation of the Gpi16 protein carboxy-terminal transmembrane domain, part of the conserved GPI transamidase domain.