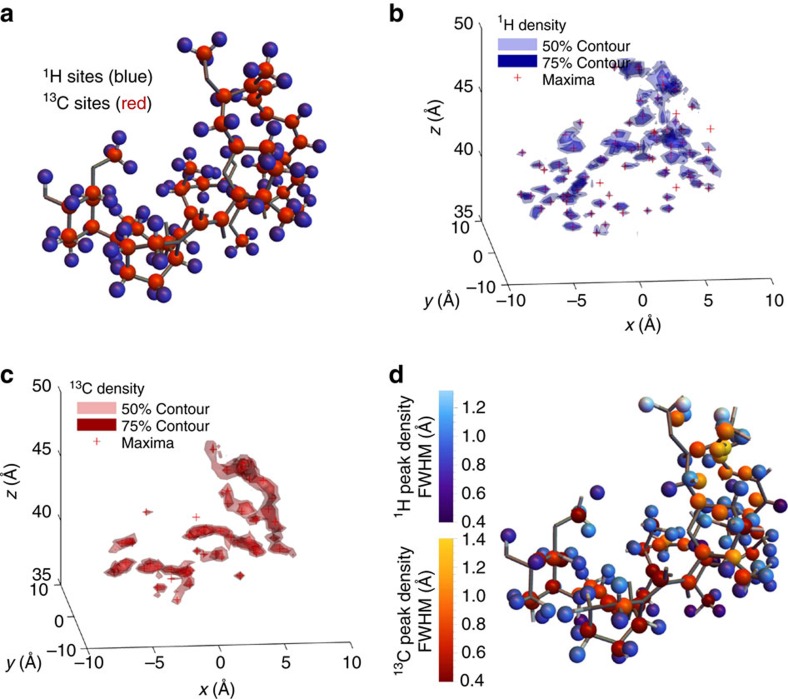
Figure 5. Simulation of the nano-MRI protocol carried out on a single rapamycin molecule (C51H79NO13).
(a) Hydrogen and carbon substructure of the rapamycin molecule. Sampling was performed over 150 slices for each of 11 × 24 latitude and longitude orientations of B0 with MS=2000 measurements per slice and sequence length t=8 ms. (b,c) Extracted 1H and 13C nuclear densities, respectively. The 3D density is represented through 50% and 75% contours that reveal subthreshold features. Atomic positions are identified corresponding to local maxima in the 3D density that are >90% or greater in relative terms (shown as crosses in (c,d)). The uncertainty in position (effective resolution) is estimated from the average of the (x,y,z) FWHM cross-sections of the 3D density peaks. Peaks closer to the probe (smaller z values) are narrower due to stronger gradient coupling to the probe, while isolated peaks are further localized due to a greater relative contrast in the numerical inversion procedure. (d) Reconstruction of the atomic positions and associated resolution (colour scales) are shown for 1H (blue colour scale) and 13C (red colour scale). The sphere size corresponds to the average statistical deviation in comparison to the exact coordinates of Δr=0.5 Å (superimposed on the data is the exact skeletal structure from a.