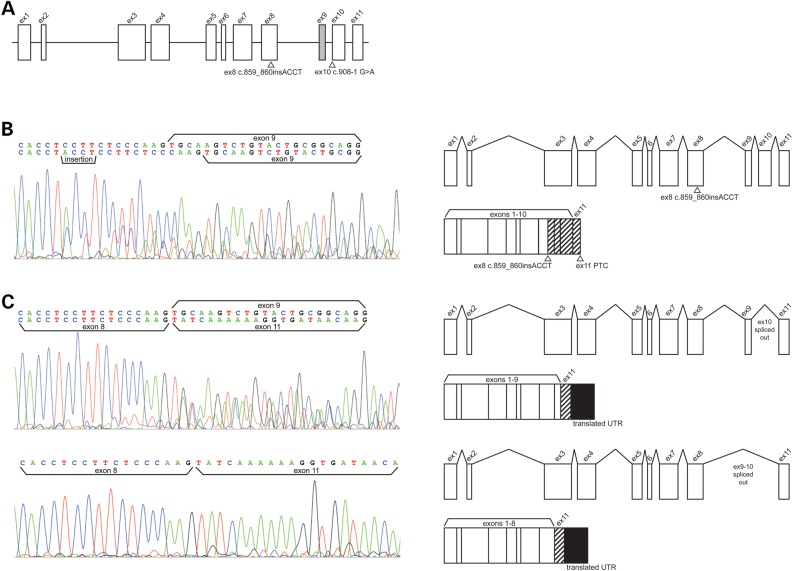
Figure 4.
Effect of CA12 variants upon RNA processing in nasal epithelial cells from proband A. (A) Exon and intron structure of CA12 with locations of proband A variants identified by NGS. Rectangles represent exons and the lines through the center of the rectangles represent the genomic axis. Variants are indicated with triangles and HGVS cDNA names. The gray rectangle indicating exon 9 is spliced out in an alternative CA12 transcript whose function and tissue distribution is unknown. (B) (Left) Electropherogram of Sanger sequencing of cDNA reverse transcribed from RNA extracted from proband A cultured nasal epithelial. Sequencing detected a heterozygous insertion variant c.859_860insACCT on a transcript retaining alternatively spliced exon 9. A second transcript detected by sequencing does not bear the insertion, retains exon 9, and is consistent with transcript lacking exon 10 as a result of the in trans variant c.908-1 G>A. Presence of this second transcript in this sequencing reaction is likely due to imperfect isolation of the similarly sized transcripts by gel purification as transcript bearing the insertion is only 89 bp longer than transcripts lacking exon 10 only. (Right) Gene models depict RNA processing of the insertion variant and the predicted gene product. The insertion variant causes a frameshift starting in exon 8, a premature termination codon in exon 11, and was predicted to generate a misfolded protein targeted by ERAD. Hashed rectangles indicate an altered exonic reading frame. (C) (Left) Electropherogram of Sanger sequencing detecting transcript missing exons 9 and 10, and a second transcript missing exon 10 only, due to heterozygous splice acceptor variant c.908-1 G>A. Imperfect isolation of transcripts in this reaction is due to alternatively spliced exon 9 which is only 33 bp long. (Right) Gene models depict the two processed RNAs and protein products lacking exon 10 observed by RT-PCR: one with exon 9 alternatively spliced out and one retaining exon 9. This variant is predicted to cause skipping of exon 10, a frameshift resulting in read-through of the native stop codon, translation of 267 nucleotides from the 3′ UTR, and a misfolded protein targeted by ERAD. Hashed rectangles indicate an altered exonic reading frame and filled rectangles indicate translation of the 3′ UTR.