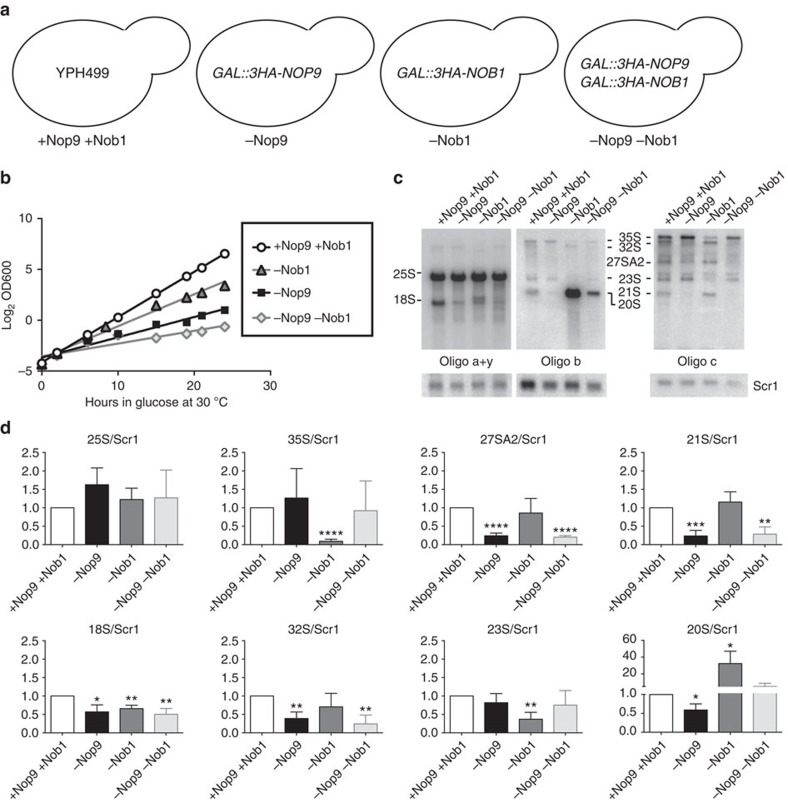
Figure 5. Probing the in vivo function of Nop9.
(a) Schematic drawings of the yeast strains used for testing the effects of Nop9 and/or Nob1 depletion. Chromosomal Nop9 and/or Nob1 expression was placed under the control of the inducible GAL promoter and sequences encoding triple-HA tags were inserted. (b) Depletion of Nop9 and/or Nob1 expression impairs growth. Representative growth curves of yeast strains grown at 30 °C for 24 h in glucose to deplete endogenous Nop9 and/or Nob1 expression. Three biological replicates were conducted. (c) Northern blot analysis of Nop9/Nob1 double depletion indicates restoration of normal 20S pre-rRNA levels. Representative northern blots detecting precursor and mature rRNA in total RNA from parental YPH499 (+Nop9 +Nob1) or yeast depleted of Nop9 (−Nop9), Nob1 (−Nob1) or both Nop9 and Nob1 (−Nop9 −Nob1) in glucose for 24 h at 30 °C. The pre-rRNA intermediates and mature rRNAs were detected using a series of oligonucleotide probes, a, b, c and y, that are indicated in Fig. 1a. Oligos a+y were used to detect 25S and 18S rRNAs, oligo b was used to detect the 20S pre-rRNA and oligo c was used to detect the 35S, 32S, 27SA2, 23S and 21S pre-rRNAs. An oligo complementary to Scr1 was used as a loading control. (d) Quantitation of replicates of the northern blots presented in c. The intensities of the mature rRNAs and pre-rRNA intermediates relative to Scr1, the loading control, were plotted. Bar graphs created in GraphPad PRISM plot the mean intensities calculated from three biological replicate experiments with error bars representing the s.e.m. The significance of the levels of mature and pre-rRNAs in −Nop9, −Nob1 or −Nop9 −Nob1 compared with +Nop9 +Nob1 YPH499 was assessed by an unpaired, two-sided t-test, and P values are indicated (*P≤0.05, **P≤0.01, ***P≤0.001 and ****P≤0.0001 nonsignificant differences have P values >0.05). The analysis of the 35S, 32S and 23S pre-rRNAs using oligo c is shown; however, similar results are obtained when analysing the blot probed with oligo b.