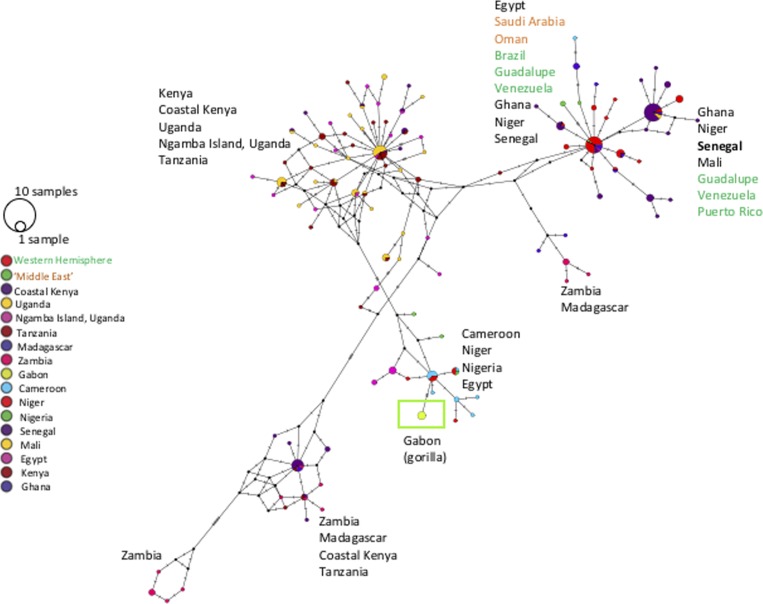
Figure 2.
Minimum spanning network inferred from Schistosoma mansoni cytochrome c oxidase subunit 1 (COI) sequences created in POPART.11 Eleven input sequences were defined by unique haplotype, identical haplotypes collapsed into a single haplotype. We started with 253 isolates of COI (311 base pairs [bp]) S. mansoni (from humans, Biomphalaria, chimps) that was transformed into a haplotype input file and had 111 unique haplotypes defined by 81 bp of unique nucleotide sites. In the key, the circle size is shown for one sample and 10 samples. Also, colored circles defined by ‘traitlabels’ in POPART represent country of origin(s) of the sequence except where some were collapsed (Western Hemisphere = Brazil, Guadalupe, Venezuela, Puerto Rico; Middle East = Saudi Arabia, Oman). For ease of visualization, country of origin is labeled next to the major clusters of those haplotypes (text color matches key). The samples of S. mansoni from gorillas in Gabon are highlighted by a green circle and box outline. The S. mansoni samples from previous reports of chimps on Ngamba Island, Uganda, did not form a unique cluster, nor did it cluster with the samples from Gabon.